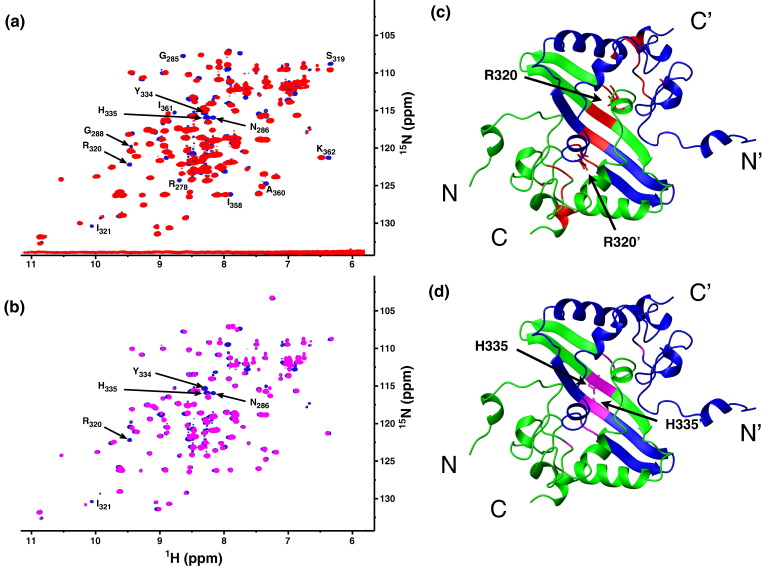
Fig. 9.
Structure perturbation of R320A and H335A mutants. (a) Overlay of 15N-edited HSQC spectra from the wild-type CTD (blue) and the R320A mutant (red). Affected resonances are identified by their respective residue types and numbers in the wild-type protein. (b) Same as in (a), but with the wild-type CTD (blue) and the H335A mutant (magenta). (c) Mapping of residues affected by the R320A mutation (red) in the solution structure of the SARS-CoV NP CTD dimer. The side chains of R320 are shown in a neon representation. (d) Same as in (c), but showing the residues affected by the H335A mutation (in magenta). The side chain of H335 is also shown.