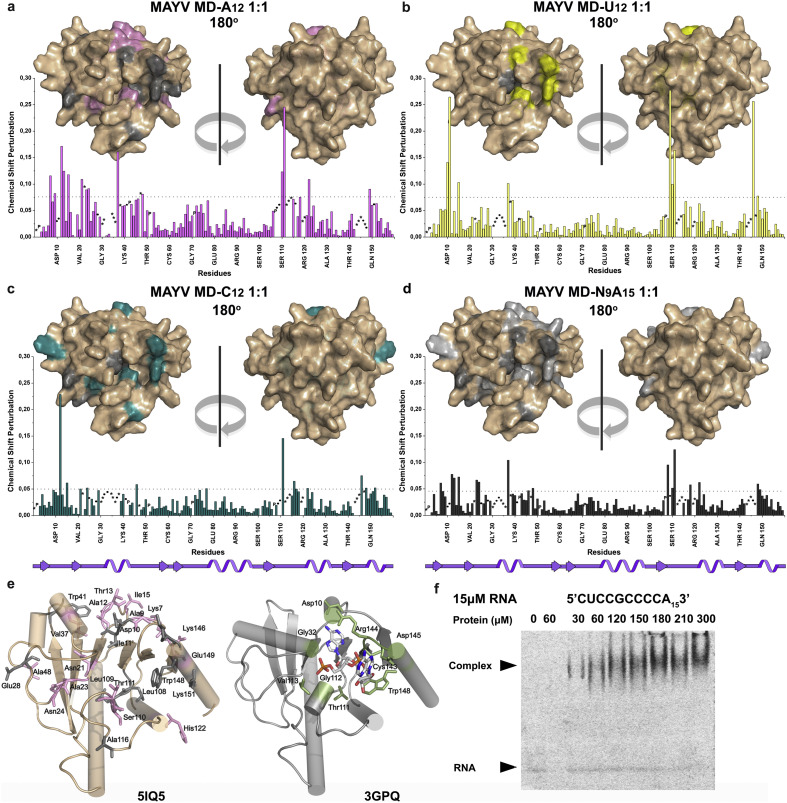
Fig. 4.
(a–d) Plots of combined amide chemical shift changes, Δδ, induced by the binding of RNA oligos to MAYV MD versus amino acid sequence at a 1:1 molar ratio of protein:RNA oligo (pink-A12, light yellow-U12, dark cyan-C12, gray-N9A15). In each plot, the dotted line indicates the applied threshold of chemical shift perturbation calculated as average value plus 1 SD. The surface representations above each plot show the amino acids exhibiting CSP values above the threshold upon binding of the respective RNA oligo to MAYV MD (color code: pink, A12; light yellow, U12; dark cyan, C12; gray, N9A15) and those with broadened amide cross peaks (in dark gray). An asterisk indicates an unassigned residue of apo-MD, p indicates a proline residue and a dot indicates a residue whose HN resonance could not be detected at each titration step. (e) Left: The amino acids of MAYV MD that are critical for binding of the A12 oligo were mapped on its solution structure using only the amino acids with CSPs above the threshold (shown as pink sticks) and those, whose signals broadened beyond detection during the titration (in gray). Right: Representation of the residues (green sticks) that bind the A3 molecule in the CHIKV MD-A3 complex (PDB 3GPQ). (f) Gel shift assay (EMSA) probing the interaction of MAYV MD with the 24mer 5’CUCCGCCCCA153’ (= N9A15). The protein concentrations used are indicated above the gel.