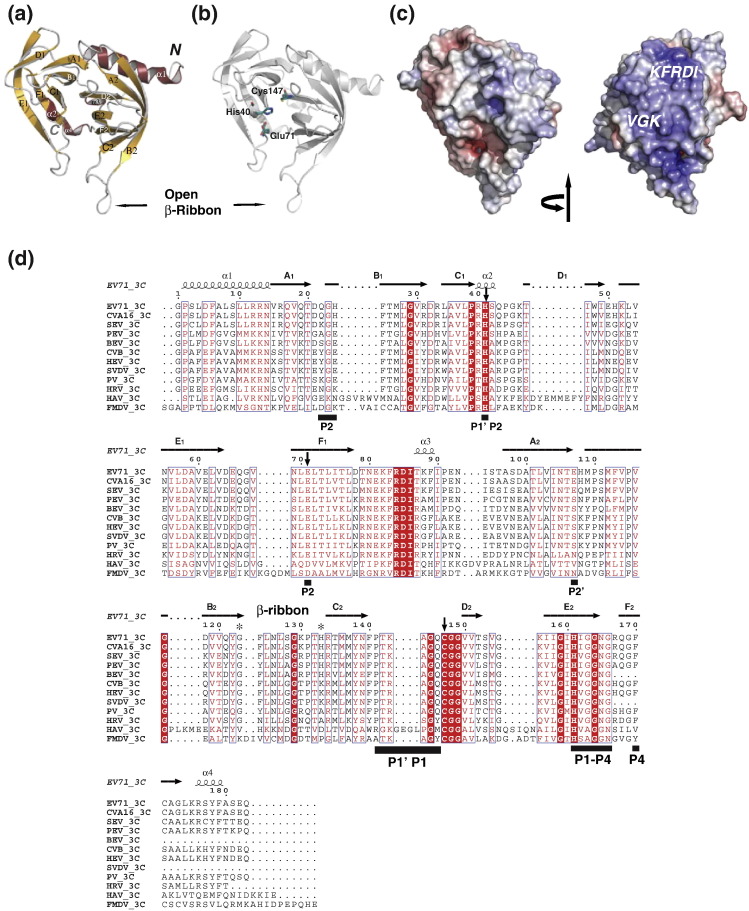
Fig. 1.
Structure of EV71 3Cpro. (a) Ribbon model of EV71 3Cpro with annotated secondary structures. The position of β-ribbon is indicated with black arrow. (b) Ribbon model of EV71 3Cpro (gray) with side chains of catalytic triad, His40, Glu71 and Cys147 (highlighted in stick presentation with deep teal for carbons). The position of β-ribbon is indicated. ⁎There was no electron density for the side chain of residue Glu71. (c) Electrostatic surface potential (ranging from blue = 20 kT/e to red = − 20 kT/e), displayed in two different views (left, “standard view” used in all other figures; right, 180° rotation around the vertical axis). Positions of the conserved RNA binding motifs “KFRDI” and “VGK” are indicated. (d) Structure-based multiple-sequence alignment of 3Cpro of different picornaviruses. The secondary structure is shown on top. Invariant residues in 3Cpro are highlighted with red background; conserved residues are shown in red font. The position of β-ribbon is indicated on the top of the sequences. Hinge residues Gly123 and His133 are indicated by asterisks. Black bars at the bottom of the sequence indicate residues directly involve in substrate binding (based on superimposition with HRV 3C–AG7088 co-structure; PDB code: 1CQQ).