Graphical abstract
.
Abbreviations: RDT, rapid diagnostic test; Pf, Plasmodium falciparum; Pv, Plasmodium vivax; NTC, no template control
Keywords: Malaria, Loop mediated isothermal amplification, Tube scanner, DNA extraction, Real-time detection, Resource limited setting
Highlights
-
•
Improved LAMP assay for the diagnosis of Malaria in a resource limited settings.
-
•
We developed a simple platform for the diagnosis of malaria by genus- and species-specific LAMP and successfully used a simple DNA extraction method.
-
•
LAMP-Tube scanner method is capable of detecting the two most common species of malaria parasites within 30 min and it’s proved to be sensitive, reliable and feasible.
-
•
Analogous results were obtained for LAMP-Tube scanner versus traditional microscopic method and LAMP-Thermocycler.
Abstract
We attempted to improve the loop-mediated isothermal amplification (LAMP) method for malaria diagnosis by using a simple DNA extraction procedure, and a portable device performing both the amplification and detection of LAMP in one platform. Additionally, the device served as a heating block for the DNA preparation. We refer this method as LAMP-Tube scanner, and evaluated using 209 microscopically positive malaria samples and compared them to RDTs and LAMP-Thermocycler. Two most common human infecting Plasmodium species were detected. The LAMP-Tube scanner method is found to be simple and allowed real-time detection of DNA amplification. The time to amplification varied but was closely less than 60 min. Sensitivity and specificity of LAMP-Tube scanner in detecting Plasmodium falciparum were 95% and 93.3%, compared to microscopy and 98.3% and 100% respectively, compared to standard LAMP-Thermocycler. In addition, it showed a detection limit of 10 and 40 copies of the parasitemia for Plasmodium vivax and P. falciparum. Accordingly, in comparison to the results obtained by microscopy, the LAMP-Tube scanner had a less divergence in sensitivity and specificity, and yielded results similar to those of LAMP-Thermocycler. This method has the great potential as a field usable molecular tool for the diagnosis of malaria and is an alternative to conventional PCR-based diagnostic methods for field use.
1. Introduction
Malaria is one of the most important tropical infectious diseases. About 350–500 million clinical episodes of malaria occur each year, and the disease is responsible for more than 1 million deaths annually (WHO, 2008). The range of areas inhabited by malaria-carrying mosquitoes is expanding due to global climate change (Patz and Olson, 2006). Thus, there is a chance of recovery of malaria, not only as an outcome of the increasing number of imported cases, but also as an effect of the rise of habitats apt for malaria-carrying mosquitoes. Therefore, the hasty diagnosis of this disease is extremely important.
For the past 100 years, malaria has been diagnosed by microscopic examination of Giemsa, Wright, or Field stained blood films (Bruce-Chwatt, 1987, Warhurs and Williams, 1996, White and Silamut, 1989). However, it is well documented that microscopy has limitations: it is time-consuming, and misdiagnosis of the infecting species is common if the microscopist lacks experience and/or when the parasitemia is low (Kain et al., 1998, Milne et al., 1994, Singh et al., 1999, Snounou et al., 1993a). More recently, serological diagnostic methods and new rapid diagnostic tests have become available, which are most commonly based on the detection of parasite antigens such as histidine-rich protein (HRP)-2 and lactate dehydrogenase (LDH) in an easy-to-use dipstick or a lateral flow format (Beadle et al., 1994, Makler et al., 1998). The advantages offered by these methods, such as a result can obtain within half an hour by unskilled technicians, are tempered by a few limitations (Moody, 2002). RDT methods do not offer improved sensitivity over microscopy, the sensitivity decreases as parasitemias fall below 100 parasites/μL (Mills et al., 1999). False positive results are observed, particularly after treatment, as the parasite antigens detected can remain in the circulation following parasite clearance. Moreover, the majority of the commercial RDTs detect HRP-2, which is only expressed by P. falciparum; however, not by other species and therefore this test offers specific diagnosis of falciparum malaria. Most of the non-HRP-2 based tests (LDH and aldolase) are usually pan-species test that allows for the speciation of P. falciparum and/or non-falciparum species when used in combination with HRP-2 based tests.
Several PCR based assays have also been developed for the detection and identification of malaria parasites. Most often based on genus- or species-specific sequences of the parasites rRNA gene (Singh et al., 1999, Kawamoto et al., 1996, Snounou et al., 1993b). PCR-based assays have various advantages over microscopy and RDTs: they are highly specific and sensitive (Kain et al., 1998, Snounou et al., 1993b, Zhong and Kain, 1999), and as few as five parasites per microliter of blood can be detected (Makler et al., 1998). Regrettably, the current PCR-based methods are beyond the capacity of most malaria-endemic countries as they need expensive inspections, sophisticated laboratory and training that make these techniques expensive and practically challenging to implement in the field or resource limited settings. The recently developed loop-mediated isothermal amplification method is a relatively simple and field-adaptable technique (Notomi et al., 2000). This method does not require a thermocycler or sophisticated training. It has the potential to be used as a molecular diagnostic tool for point-of-care (POC) testing in both developing and developed countries. LAMP has been used for the detection of several infectious diseases such as severe acute respiratory syndrome (Hong et al., 2004), West Nile virus (Parida et al., 2004), avian influenza virus (Imai et al., 2006), norovirus (Fukuda et al., 2006) and Legionella bacteria (Annaka et al., 2003).
Lately, the LAMP assay for malaria parasites has already been reported by Poon et al., 2006, Han et al., 2007, Paris et al., 2007, Yamamura et al., 2009. However, LAMP assays can be used in the resource limited areas, but there is a need for electricity to power equipment such as thermocycler, water bath, centrifuge etc., second, they require a complex procedure for the sample preparation. Although highly specific and sensitive, the time and resources needed are restricting it from being used in tribal areas. Therefore, there is a requirement for a minimal DNA preparation and field-usable instrument that can allow a faster and objective readout for the diagnosis of malaria in resource-limited settings. Here, we report a simple DNA extraction procedure and the use of a mini portable device in which both amplification and real-time detection units are combined into a single unit for LAMP assay. We regard this method as LAMP-Tube scanner.
2. Materials and methods
2.1. Study site and sample collection
Clinically related work in this study was performed at Genomix Molecular Diagnostics Pvt. Ltd., (Hyderabad, India) and the portable device connected work carried out at QIAGEN Lake Constance GmbH (formerly ESE-GmbH, Stockach, Germany). For field diagnosis of malaria, 52 febrile patients were analyzed on-site by LAMP-Tube scanner method. Microscopy, LAMP-Thermocycler and RDTs were performed off-site for the same samples. For the laboratory detection of malaria, a total of 128 blood samples that were positive for P. falciparum and 40 samples that were positive for P. vivax and 15 samples that were negative for malaria parasites by microscopy and RDTs collected from the clinical diagnostic centers and medical institutes throughout the Andhra Pradesh, southern province of India, where P. falciparum and P. vivax are the common most parasites transmitted. Patients samples previously diagnosed with malaria and treated with antimalarials were excluded. All blood samples and samples saved as blood spots on filter paper were stored at −20 °C. Purified P. falciparum DNA (3D7) was provided by Dr. Udhayakumar Venkatachalam (CDC, Atlanta, USA).
2.2. Microscopy
Thick and thin blood films were stained according to the method described by Field (White and Silamut, 1989) and examined by an experienced microscopist. Parasitemia was assessed either per 1000 erythrocytes in the thin film or at low parasitemias per 200 white blood cells in the thick film. In case of a putative negative film, 500 WBC were assessed to confirm the absence of parasites. The number of parasites/μL was determined by counting the total number of RBCs/μL.
2.3. Rapid diagnostic test
The blood samples were also analyzed with the Malaria test kits (Genomix Molecular Diagnostics Pvt. Ltd., Hyderabad, India), based on immunological detection of the P. falciparum specific histidine-rich protein 2 (PfHRP2) and P. vivax specific lactate dehydrogenase (pLDH). The assays were performed in parallel according to the manufacturer’s instructions using a drop of EDTA anticoagulated whole blood and the results were observed after 15 min by the naked eye.
2.4. Processing of samples and DNA extraction for LAMP
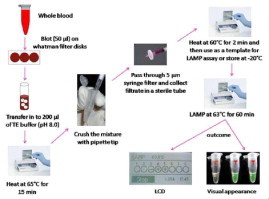
The DNA template used for LAMP-Thermocycler (LAMP performed using a thermocycler, Bio-Rad, USA) was prepared from 200 μL of whole blood using the QIAamp DNA Blood Mini kit (QIAGEN, Hilden). For the LAMP-Tube scanner (LAMP performed using a tube scanner, QIAGEN Lake Constance GmbH, Germany), a second simple and cheap method of DNA extraction was used and described in this work (Fig. 5 ). Briefly, 50 μL of human blood was directly blotted onto Whatman filter paper disks and air-dried at room temperature for 5 min. Blotted disks were heated in 200 μL of TE buffer (pH 8.0) at 65 °C for 15 min followed by crushing the disks and filtered through 5 μm syringe filter. The filtrate was heated at 60 °C for 2 min before proceeding to LAMP. Two microliters of the resulting filtrate was used as a template for the LAMP assay and the remaining sample was stored at −20 °C until further analysis. To test the limits of detection of the LAMP-Tube scanner, DNA from the samples was diluted from 1000 p/μL to 1 p/μL. Sixty samples confirmed to be P. falciparum positive by microscopy and 30 samples known to be negative for P. falciparum were used to assess the sensitivity and specificity of the LAMP-Tube scanner. Non-malaria infected human DNA was used as a control.
Fig. 5.
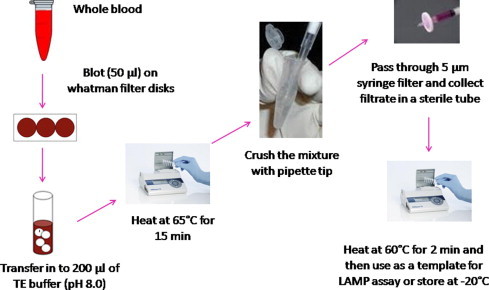
Schematic representation of the DNA extraction method described and used in this study for the Malaria LAMP-Tube scanner. Detailed process explanation is given in Section 2.4.
2.5. LAMP assay
The LAMP reaction was performed with the Loopamp DNA Amplification Kit (Eiken Chemicals Co., Tokyo, Japan) following the manufacturer’s instructions. Genus- and species-specific primers, as described by Han et al., 2007; were used to amplify the gene coding for the 18s ribosomal RNA. The reaction consisted of 6 μL of primers (FIP and BIP 1.6 μM, Loop-F and Loop-B 0.8 μM, F3 and B3 0.2 μM), 12.5 μL of reaction buffer, 1 μL Bst DNA polymerase, 1 μL of fluorescent detection reagent (Eiken Chemicals Co., Tokyo, Japan), 2 μL template DNA, and distilled water to a total volume of 25 μL. To test the avail of an in-house reaction buffer, pilot experiments were performed in a 12.5 μL total volume containing a 2× in-house reaction buffer (40 mM Tris–HCl, 20 mM NH4SO4, 20 mM KCL, 16 mM MgSO4, 0.2% Triton X-100, 1.6 M glycine betaine, 2.8 mM dNTP’s each), 1 μL of 0.5 mM calcein and 8 units of Bst polymerase (New England Biolabs, MA, USA). DNA amplification was carried out at 63 °C for 90 min using both the thermocycler and tube scanner. A unique real-time amplification plot obtained using the LAMP-Tube scanner is shown in Fig. 1 B, which was set to collect fluorescence signals at 20-s intervals. In the plot, the Y-axis indicates the fluorescence units in milli-volts (mV) and X-axis demonstrates the time in minutes. Amplification of P. falciparum DNA produced a sigmoid shaped amplification curve while the control tube (no DNA) had no measurable change in fluorescence intensity denoted by a flat line in the plot.
Fig. 1.
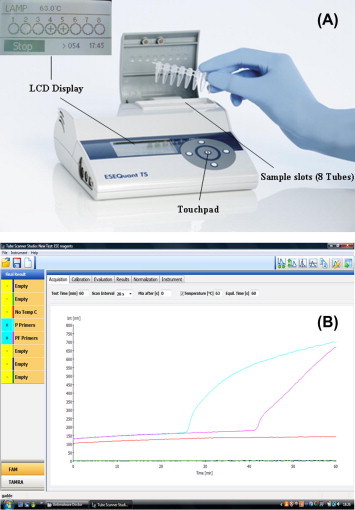
Description of the LAMP-Tube scanner. The tube scanner assembled with temperature settings to amplify DNA isothermally and spectral tools to distinguish amplified product using fluorescence is shown (A). The tube scanner can hold 8, 200 μL PCR tubes and is assembled with an LCD screen through which positive or negative results can be seen. If the tube scanner is connected to a computer with the suitable software, the results are obtained in real-time as shown in (B). The fluorescence units are shown on the Y-axis and time to amplification on the X-axis. Amplification curves are observed (curved lines, Plasmodium genus specific primers – blue lane, Plasmodium falciparum species specific primers – magenta color) in case of positive samples. No amplification curves (flat line, orange color) indicated a negative sample. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.6. Analysis of the LAMP products
The LAMP reaction causes turbidity in the reaction tube corresponding to the amount of amplified DNA. Hence, the turbidity was observed with the naked eye. To confirm the sensitivity and the possibility of real-time LAMP quantification of Plasmodium parasites, fluorescence was monitored by the tube scanner. For further validation, all reactions were analyzed by 1.5% agarose gel electrophoresis and positive results were defined by the appearance of typical ladder bands of various sizes (Han et al., 2007).
2.7. About the portable device
The portable equipment (ESE Quant tube scanner) used for this work was developed by the QIAGEN Lake Constance GmbH, formerly ESE-GmbH, Stockach, Germany (Fig. 1A). This device weighs about 2.2lbs having the dimensions 74 mm × 178 mm × 188 mm (H × W × D). It has an eight tube holder heating block with adaptable temperature settings and spectral tools to distinguish amplified products using fluorescence array. The device is totally carriable and can be operated outdoors with a Li-Ion rechargeable power pack. A small LCD monitor is built-into display the results as positive or negative (+/−) without the need of a computer. Nevertheless, the device can also be used together with a computer to create real-time amplification plots as the reaction progresses.
2.8. Statistics
The sensitivity and specificity of the LAMP-Tube scanner were determined using microscopy, RDTs and LAMP-Thermocycler as reference methods. Sensitivity was calculated as the (number of true positives)/(number of true positives + number of false negatives) × 100, and specificity was calculated as the (number of true negatives)/(number of true negatives + number of false positives) × 100.
3. Results
3.1. Malaria-LAMP-Tube scanner
We could detect the two most common species of human malaria parasites (P. falciparum and P. vivax) within 30 min using the LAMP-Tube scanner (Fig. 2 ). No amplification was observed with the non-malaria infected human DNA control (Fig. 2). Results were also interpreted by the visual observation (Fig. 3 ) and agarose gel electrophoresis (Fig. 4 ). A total of 168 [P. falciparum (128) and P. vivax (40)] microscopically positive samples were tested in the laboratory. LAMP-Thermocycler, which was used as a reference method, was positive for P. falciparum in 124 samples and for P. vivax in 38 samples (Table 1 ), whereas the LAMP-Tube scanner was positive for 123 and 38 samples, respectively. RDTs, which were used as a second reference method, were positive for 116 and 34 samples. Out of 52 suspicious malaria samples tested in the field, 38 samples were positive by LAMP-Tube scanner (Table 1), for LAMP-Thermocycler, 37 were positive, for RTDs, 31 were positive and 41 were positive by microscopy. In comparison to referential methods, there were no significant differences observed by LAMP-Tube scanner, despite more false- negative results were obtained by RTDs.
Fig. 2.
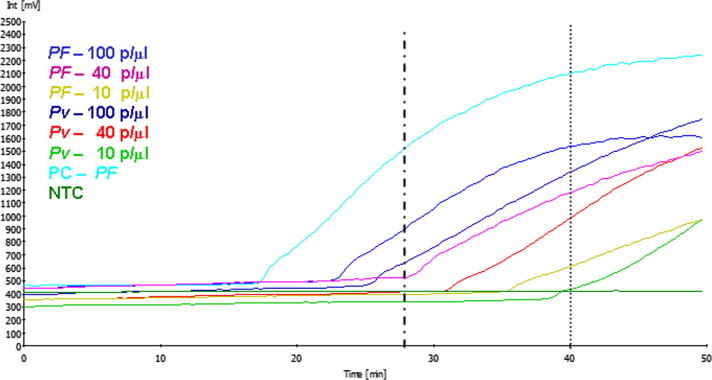
Amplification of the two most human-infecting Plasmodium species using the LAMP-Tube scanner method. Plasmodium species-specific primers were used to amplify the 18s ribosomal RNA gene in P. falciparum and P. vivax parasites. Amplification curves (positive) were observed for the two species (100 parasitemia/μL) within 30 min (vertical dotted line), and with the low parasitemia (40 & 10 p/μl), amplification curves were seen in 40 min (vertical dotted line). No amplification was seen having the negative control or in the no template control (NTC). As a positive control (PC), purified Plasmodium falciparum DNA (3D7) was amplified in 18 min.
Fig. 3.

Visual appearance of LAMP-Tube scanner reactions using calcein (fluorescent dye). The color changes from orange (negative reaction) to yellowish green (positive reaction).
Fig. 4.
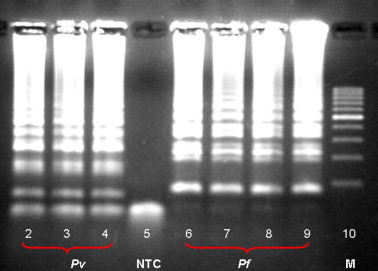
Agarose gel electrophoresis of the malaria LAMP-Tube scanner products. One microliter aliquots of the LAMP products were run on a 1.5% agarose gel. A clear ladder-like pattern was observed in the positive reactions [lanes 2,3,4 (pv) & 6,7,8,9 (pf)], whereas no ladder pattern was seen in the negative control (lane 5). Lane M, represents the DNA ladder marker.
Table 1.
Comparison of microscopy, RTDs, LAMP-Thermocycler, and LAMP-Tube scanner for malaria parasite detection and species identification.
| Laboratory detection | ||||||
|---|---|---|---|---|---|---|
| Total no of samples |
RDTs |
LAMP-Thermocyclera |
LAMP-Tube scannerb |
|||
| (n = 168)/microscopy | Positive | Negative | Positive | Negative | Positive | Negative |
| P. falciparum (128) | 116 | 12 | 124 | 4 | 123 | 5 |
| P. vivax (40) | 34 | 6 | 38 | 2 | 38 | 2 |
| Control (15) | 1 | 14 | 0 | 14 | 0 | 14 |
| Field detection | ||||||
| Total No of samples (n = 52)/Lamp-Tube scannerb | Microscopy | |||||
| Positive | Negative | |||||
| P. falciparum (25) | 21 | 4 | 25 | 0 | 25 | 0 |
| P. vivax (13) | 10 | 3 | 12 | 1 | 13 | 0 |
| Negative (14) | 0 | 14 | 0 | 14 | 3⁎ | 11 |
DNA extraction was performed using the QIAGEN kit.
DNA extraction was performed using a simple method explained in this study (Fig. 5).
2 – Pf positive and 1 – Pv positive.
3.2. Detection limits of Plasmodium species-specific LAMP-Tube scanner
The detection limits of the LAMP-Tube scanner were assessed using DNA obtained from P. falciparum and P. vivax. DNA was diluted from 1000 p/μL to 1 p/μL. The limits of detection of the LAMP-Tube scanner varied between 10–100 p/μL for both species (Table 2 ). This method needed at least 10–100 p/μL for the detection of P. falciparum. A minimum of 10–40 p/μL was required for the detection of P. vivax (Table 2). The LAMP-Thermocycler detected up to 40 p/μL for the two species (data not shown). The time required for amplification varied between 15–60 min. Additional time to amplification was needed for samples possessing lower parasite densities even though no clear correlation was observed between time to amplification and parasite densities.
Table 2.
Detection limits of the LAMP-Tube scanner method tested using various dilutions of P. falciparum and P. vivax.
| Lowest conc. detected (p/μl) | P. falciparum | P. vivax |
|---|---|---|
| Assay # 1 | 10 | 10 |
| Assay # 2 | 40 | 10 |
| Assay # 3 | 40 | 40 |
| Assay # 4 | 100 | 10 |
| Assay # 5 | 40 | 40 |
| Assay # 6 | 100 | 40 |
3.3. Clinical sensitivity and specificity
Clinical samples with median parasitemia of 1000 p/μL (parasitemia per microliter), were used to test the utility of this platform for the diagnosis of field samples. A schematic workflow of sample preparation, amplification and detection, is shown in graphical abstract. The sensitivity and specificity of the LAMP-Tube scanner method compared to microscopy, LAMP-Thermocycler and RTDs were shown in Table 3 . Among the 60 microscopically positive samples (P. falciparum) tested, 58 samples were shown to be positive by the LAMP-Thermocycler and 57 positive by LAMP-Tube scanner. Twenty eight out of the 30 microscopically negative samples were confirmed to be negative by these two methods. Overall, both LAMP-Tube scanner and LAMP-Thermocycler yielded results very similar to those of microscopy (Table 3). The LAMP-Tube scanner showed 98.3% sensitivity and 100% specificity when compared to LAMP-Thermocycler. RDTs produced more false-positive results than the two LAMP methods.
Table 3.
Sensitivity and specificity of the LAMP-Tube scanner, LAMP-Thermocycler and RDTs compared to microscopy.
| Microscopy (n) | RDTs |
LAMP-Thermocycler |
LAMP-Tube scanner |
|||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| Positive (60) | 52 | 8 | 58 | 2 | 57 | 3 |
| Negative (30) | 5 | 25 | 2 | 28 | 2 | 28 |
| Sensitivity (%) | 86.7 | 96.7 | 95.0 | |||
| Specificity (%) | 83.3 | 93.3 | 93.3 | |||
3.4. Commercial vs. in-house buffer for the LAMP-Tube scanner
We compared a LAMP reaction buffer obtained from the commercial kit (Eiken Chemicals, Japan) to one prepared in-house for their performance in the LAMP-Tube scanner method. As depicted in Fig. 6 , both buffers were able to begin the amplification in 35 min. However, the intensity of the amplification was slightly lower when an in-house buffer was used. Overall, the qualitative results were not affected by the two buffers, as both were displayed positive for P. falciparum within 37 min (data not shown). Compared to commercially available buffer, the cost of running the LAMP-Tube scanner was manifold (about 20 times) lower when an in-house buffer was used.
Fig. 6.
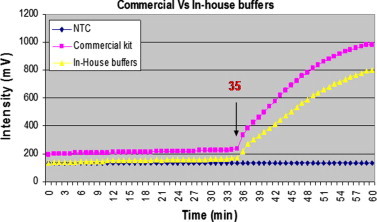
Comparison of in-house and commercial buffers for the P. falciparum LAMP-Tube scanner. More or less similar amplification curves were observed for both buffers in 35 min. No amplification was seen with no template control (NTC).
3.5. Quantification using the LAMP-Tube scanner
The purified lambda phage DNA obtained from Sigma–Aldrich (Germany), was quantified using the LAMP-Tube scanner assay. A starting DNA concentration of 100,000 pg/μL was prepared from which five 10-fold serial dilutions were prepared to a final DNA concentration of 1 pg/μL. As shown in Fig. 7 , a reasonable correlation was observed between each dilution and intersection of real-time curves except the least concentration (1 pg/μl). The intersection of real-time curves at the threshold level was chosen for the regression analysis (R-square analysis). A very good statistical correlation of 99.79% was obtained using linear regression, excluding the final DNA concentration (Fig. 7B).
Fig. 7.
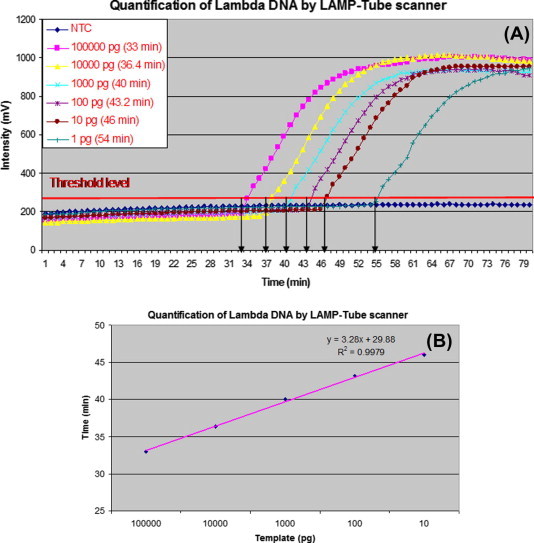
Quantification of Lambda phage DNA by the LAMP-Tube scanner. (A) Amplification curves of all the concentrations used (1–100,000 pg). (B) The statistical significance of the peak rising points just above the threshold level (excluding 1 pg concentration).
4. Discussion
The newly described LAMP technique has the feasibility of combining the accuracy of a molecular technique with the low cost and low technical requirements of the antigen-based tests. The costs for the assay are even lower than the presently available RDTs (Poon et al., 2006). LAMP has several advantages over current methods for the molecular diagnosis of disease. The Bst polymerase catalyzing the LAMP reaction is more robust to inhibition than Taq polymerase. It is, therefore, possible to use a modest, rapid and inexpensive regime for sample preparation, unlike those required for standard PCR. Furthermore, the isothermal nature of the LAMP reaction eliminates the need for expensive thermal cyclers. However, the equipment and power requirements for the DNA extraction and LAMP reaction still be a major consideration for many diagnostic facilities. This is an obvious drawback for resource-poor locations, where malaria is endemic. Some studies have clearly demonstrated that LAMP can be used for malaria diagnosis (Poon et al., 2006, Han et al., 2007, Paris et al., 2007, Yamamura et al., 2009, Zheng et al., 2002). In this study, we have developed a simpler method (LAMP-Tube scanner) for the diagnosis of clinical malaria, including species identification, based on the LAMP. We integrated the amplification and detection phases of LAMP into one portable and simple-to-use platform in an attempt to permit this method to be easily usable in field settings.
Results from the LAMP-Tube scanner (Table 3) were comparable to the previously reported malaria LAMP assays (Poon et al., 2006, Han et al., 2007, Paris et al., 2007, Yamamura et al., 2009, Zheng et al., 2002) exhibiting acceptable sensitivity and specificity profiles when compared to microscopy, RDTs and nested PCR. The reported sensitivity and specificities by these studies ranged from 73.1% to 98.3% and 85.7% to 100%, respectively, using microscopy as a reference standard. The use of various reference tests, and differences in the parasite densities of the samples used in the various studies, undoubtedly influence the sensitivities and specificities which could explain the differences observed across these studies. Out of the 155 (P. falciparum) & 54 (P. vivax) microscopically positive samples used in this study, 6 and 4 were negative by LAMP-Thermocycler and 7 and 3 by LAMP-Tube scanner. Failing to amplify these samples by the two LAMP methods was not due to low parasite density since the level of parasitemia was within the level detectable by LAMP, but most probably due to a poor quality of the DNA preparation. However, more (28) RDTs false-negative results might be due to the insufficient parasite density and/or material used to manufacture RDT strips. Additional prospective studies in different transmission settings will be needed to further evaluate the performance of the LAMP-Tube scanner method.
One of the limitations of the current study is that the LAMP-Tube scanner method was evaluated using only P. falciparum and P. vivax clinical samples. Though the LAMP method can be used for the diagnosis of all four human-infecting malaria parasites (Han et al., 2007), attempts were made only for these two parasites due to clinical sample availability. However, this genus- and species-specific LAMP-Tube scanner method could be used for the detection of all human malaria parasites. It can also be used as a confirmatory test for malaria infection in place of a standard PCR-based assay. The observation that the lowest level of parasitemia required to detect P. falciparum was higher than that needed for the P. vivax tested may be due to variations in the DNA content at different stages of the parasite. These limits of detection of species-specific LAMP-Tube scanner method are similar to those reported for microscopy, RDTs and LAMP-Thermocycler.
LAMP assay has previously been used to quantify plasmid DNA harboring the 18S rRNA genes of all four human malaria parasites (Han et al., 2007), but we failed to quantify P. falciparum DNA using the LAMP-Tube scanner. We observed that the time to amplification was shorter for samples with high parasite densities than with low parasite densities. A strict correlation was not observed when samples were compared between runs indicating that one cannot draw conclusions about parasitemia levels based on the amplification time. However, we quantified Lambda phage DNA using the LAMP-tube scanner, although it is not a perfect assay for quantification. We applied the average threshold time for the linear regression analysis. A very good statistically significant linear correlation was seen between all DNA concentrations except the least concentration. Possible explanations are that the longer incubation time before the detectable amplification caused by the lower template DNA concentration reduces the enzyme activity and decreases the annealing efficiency of the primers, may cause a delay in the threshold time. Further improvement is needed to determine that this method can be used for the quantification of malaria parasite.
In this study, we were able to successfully use a simple DNA extraction procedure (Fig. 5) for the malaria LAMP-Tube scanner without the help of any instruments other than the Tube scanner. Tube scanner, which is used as a heating block for DNA preparation and amplification, was also served as a fluorescence detector for the real-time amplification. This novel method yielded DNA extract that reliably detects as low as 10 p/μL and showed slightly lower efficiency compared to the Qiagen method. It is not clear whether this difference was due to poor efficiency in DNA extraction at low parasitemia level or due to other factors. The simple heat-treated method yielded DNA extract that could be used to detect as low as 40 p/μL, has already been described (Poon et al., 2006, Lucchi et al., 2010). However, the need for a water bath and centrifuge means that this method is not as convenient as LAMP-Tune scanner in the field and resource limited settings.
As shown in Table 3, both LAMP-Tube scanner and LAMP-Thermocycler yielded results very similar to those of microscopy. The cost of running the LAMP-Tube scanner method was cheaper than that of the LAMP-Thermocycler when an in-house buffer was used in place of the commercially available buffer. The performance of our in-house buffer was as good as commercial buffer, and it yielded consistently similar results. There are various significant features of the LAMP-Tube scanner that allow it to be an attractive method for field use. This includes, the fact that the tube scanner is easily carriable to remote places while thermocyclers need an established laboratory setting, battery can be used to run the tube scanner, no post-run analyses such as gel electrophoresis is required to visualize the results and finally, the LAMP-Tube scanner method is practically easier to perform than the LAMP-Thermocycler and nested PCR. The tube scanner is equivalent to the real-time turbidimeter used in some studies (Poon et al., 2006) in the sense that both are able to detect a positive sample in real-time resulting in same amplification plots. The turbidimeter measures the turbidity of the reaction mixture while the LAMP-Tube scanner measures the fluorescence units generated as the product is formed. However, the tube scanner has an added feature in which the results (+/−) can be reported on the provided LCD without the need of a computer.
The use of any diagnostic test for point-of-care and field use will lie among other things, on the fact that it is less expensive and simpler to perform without compromising its sensitivity and specificity. Development of LAMP reaction components in a lyophilized format would minimize the preparation time, limit the training required by the operator and removes the necessity to store the reaction components in a frozen form. The LAMP-Tube scanner method will be more attractive for field use if a lyophilized reagent format is achieved. Lyophilized buffer format which is under development, together with this simple DNA extraction and portable battery-powered instrument, offers a tangible route for the detection of malaria in limited resource settings. In conclusion, the LAMP-Tube scanner developed in this study can be used as a point-of-care tool for clinical diagnosis and active surveillance of malaria parasites in countries where malaria is endemic.
Acknowledgments
This work was supported by the Department of Biotechnoloty, Government of India and by the Qiagen lake Constance GmbH, Germany. We are grateful to the DBT and Mr. Klaus Haberstroh (QIAGEN), for financial and technical support.
References
- Annaka T., Yoshino M., Momoda T. Rapid and simple detection of Legionella species by LAMP, a new DNA amplification method. J. Jpn. Soc. Clin. Microbiol. 2003;13:19–25. [PubMed] [Google Scholar]
- Beadle C., Long G.W., Weiss W.R., McElroy P.D., Maret S.M., Oloo A.J., Hoffman S.L. Diagnosis of malaria by detection of P. faciparum HRP-2 antigen with a rapid dipstick antigen-capture assay. Lancet. 1994;343:564–568. doi: 10.1016/s0140-6736(94)91520-2. [DOI] [PubMed] [Google Scholar]
- Bruce-Chwatt L.J. From Laveran’s discovery to DNA probes: new trends in diagnosis of malaria. Lancet. 1987;ii:1509–1511. doi: 10.1016/s0140-6736(87)92634-1. [DOI] [PubMed] [Google Scholar]
- Fukuda S., Takao S., Kuwayana M., Shimazu Y., Miyazaki K. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2006;44:1376–1381. doi: 10.1128/JCM.44.4.1376-1381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han E.T., Watanabe R., Sattabongkot J., Khuntirat B., Sirichaisinthop J., Iriko H., Jin L., Takeo S., Tsuboi T. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 2007;45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M., Odagiri T. Development of H5-RT-LAMP system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Kain K.C., Harrington M.A., Tennyson S., Keystone J.S. Imported malaria: prospective analysis of problems in diagnosis and management. Clin. Infect. Dis. 1998;27:142–149. doi: 10.1086/514616. [DOI] [PubMed] [Google Scholar]
- Kawamoto F., Miyake H., Kaneko O., Kimura M., Dung N.T., Liu Q., Zhou M., Duc Dao L., Kawai S., Isomura S., Wataya Y. Sequence variation in the 18S rRNA gene, a target for PCR-based malaria diagnosis, in Plasmodium ovale from Southern Vietnam. J. Clin. Microbiol. 1996;34:2287–2289. doi: 10.1128/jcm.34.9.2287-2289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi N.W., Demas A., Narayanan J., Sumari D., Kabanywanyi A., Kachur S.P., Barnwell J.W., Udhayakumar V. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS ONE. 2010;5(10):1–7. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makler M.T., Piper R.C., Milhous W.K. Lactate dehydrogenase and the diagnosis of malaria. Parasitol. Today. 1998;14(9):376–377. doi: 10.1016/s0169-4758(98)01284-8. [DOI] [PubMed] [Google Scholar]
- Mills C.D., Burgess D.C.H., Taylor H.J., Kain K.C. Evaluation of a rapid and inexpensive dipstick assay for the diagnosis of Plasmodium falciparum malaria. Bull. WHO. 1999;77:553–559. [PMC free article] [PubMed] [Google Scholar]
- Milne L.M., Kyi S., Chiodini P.L., Warhurst D.C. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J. Clin. Pathol. 1994;47:740–742. doi: 10.1136/jcp.47.8.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody A. Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris D.H., Imwong M., Fiaz A.M., Hasan M., Yunus E.B., Silamut K., Lee S.J., Day N.P.J., Dondorp A.M. Loop-mediated isothermal PCR for the diagnosis of falciparum malaria. Am. J. Trop. Med. Hyg. 2007;77:972–976. [PubMed] [Google Scholar]
- Patz J.A., Olson S.H. Malaria risk and temperature: influences from global climate change and local land use practices. PNAS USA. 2006;103:5635–5636. doi: 10.1073/pnas.0601493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Ma E.H., Chan K.H., Chow L.M., Abeyewickreme W., Tangpukdee N., Yuen K.Y., Guan Y., Looareesuwan S., Peiris J.S. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chemother. 2006;52:302–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- Singh B., Bobogara A., Cox-Singh J., Snounou G., Abdullah M.S., Rahman H.A. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am. J. Trop. Med. Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- Snounou G., Viriyakosol S., Jarra W., Thaithong S., Brown K.N. Identification of the four human malarian parasite specific species in field samples by the polymerase chain reaction and detection of high prevalence of mixed infections. Mol. Biochem. Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- Snounou G., Viryakosol S., Zhu X.P., Jarra W., Pinheiro L., Rosario V.E., Thaithong S. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- Warhurs D.C., Williams J.E. Laboratory diagnosis of malaria: ACO broadsheet no.148. J. Clin. Pathol. 1996;49:533–538. doi: 10.1136/jcp.49.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J., Silamut K. Rapid diagnosis of malaria. Lancet. 1989;1:435. doi: 10.1016/s0140-6736(89)90025-1. [DOI] [PubMed] [Google Scholar]
- WHO, 2008. World Malaria Reports. World health organization, Geneva.
- Yamamura M., Makimura K., Ota Y. Evaluation of a new rapid molecular diagnosis system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification and melting curve analysis. Jpn. J. Infect. Dis. 2009;62:20–25. [PubMed] [Google Scholar]
- Zheng C., Xie P., Chen Y. Recombinant mycobacterium bovis BCG producing the circumsporozoite protein of Plasmodium falciparum FCC-1/HN strain induces strong immune responses in BALB/c mice. Parasitol. Int. 2002;51:1–7. doi: 10.1016/s1383-5769(01)00094-0. [DOI] [PubMed] [Google Scholar]
- Zhong K., Kain K.C. Evaluation of a colorimetric PCR-based assay to diagnose Plasmodium falciparum malaria in travelers. J. Clin. Microbiol. 1999;37:339–341. doi: 10.1128/jcm.37.2.339-341.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


