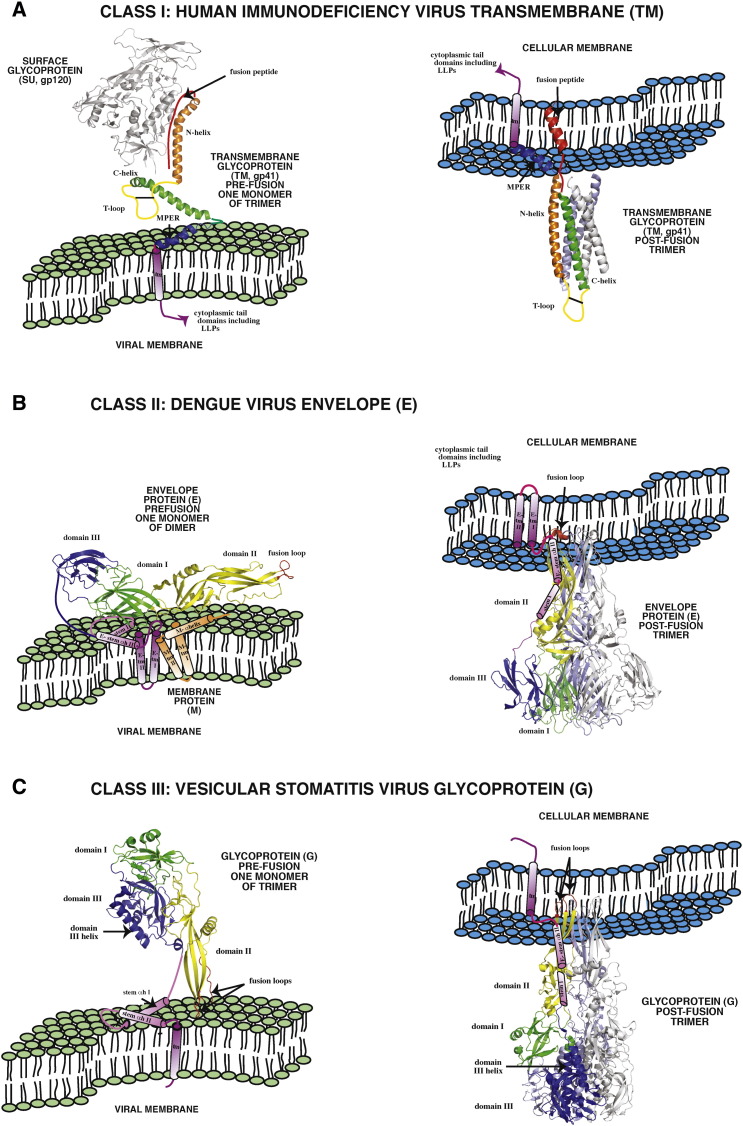
Fig. 2.
Pre- and post-fusion structures of representative viral fusion proteins of Classes I–III. The domain architectures of the three known classes of viral fusion proteins (VFP) of enveloped viruses are shown in the pre-fusion (virion) and post-fusion states. Panel A. The domain architecture of human immunodeficiency virus (family: Retroviridae) transmembrane glycoprotein (TM), a Class I VFP is indicated: red: fusion peptide, orange: amino terminal helix (aka helical region 1, HR1 or NHR), yellow: T loop containing the dicysteine bridge, green: carboxyl terminal helix (aka HR2 or CHR), blue: membrane proximal region (MPER aka aromatic domain), indigo: transmembrane domain (tm). Pre-fusion structure: protein database 4NCO, [135]. Post-fusion structure: 3F4Y, [136]. Fusion peptide in post-fusion state: 1ERF, [137]. MPER structure: 1JAU, [138]. Panel B. The domain architecture of dengue virus (family: Flaviviridae) envelope glycoprotein (E), a Class II VFP is indicated: green (domain I), yellow (domain II) and blue (domain III). The fusion loop, stem and tm helices of E are red, indigo and violet, respectively. The membrane protein (M) is depicted in orange. Pre-fusion structure:1OAN, [139]. Post-fusion structure: 4GSX, [140]. Panel C. The domain architecture of vesicular stomatitis virus (family: Rhabdoviridae) glycoprotein (G), a Class III VFP is indicated: green (domain I), yellow (domain II) and blue (domain III). The fusion loops, stem and tm helices of G are red, indigo and violet, respectively. Pre-fusion structure: 2J6JX, [141]. Post-fusion structure: 2CMZ, [142]. The left portion of Panel B and the depiction of cellular and viral membranes are modified from Fig. 4 of Zhang et al. [143].