Fig. 3.
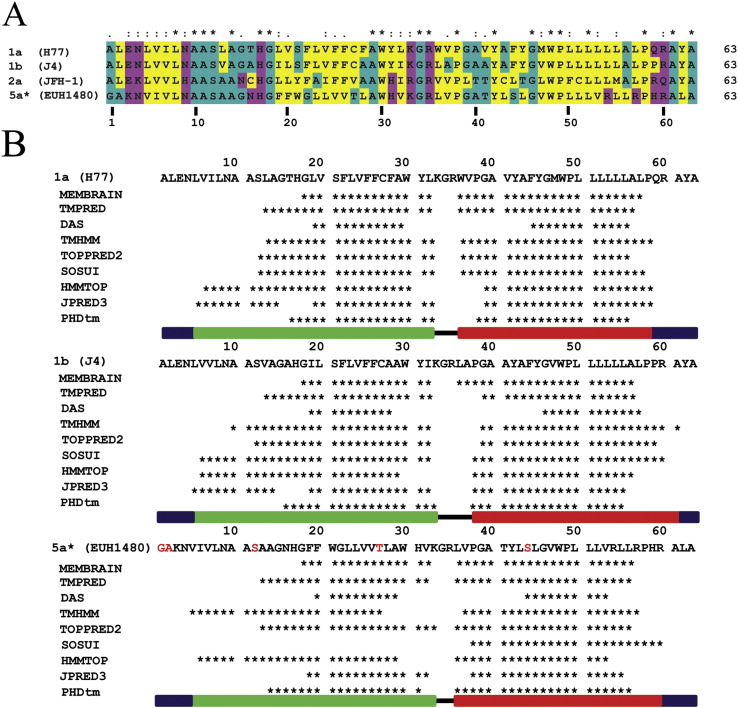
Multiple alignment (ClustalX) of the sequences of GTs 1a (H77), 1b (J4), 2a (JFH-1) and 5a (EUH1480, contains 5 mutations of unconserved amino acids (T1G, C2A, A12S, C27T, C44S marked in red)) (A). The colour scheme identifies hydrophobic amino acids in yellow, hydrophilic amino acids in magenta and neutral amino acids in cyan. The similarity index (ClustalX, top row) is as follows: ‘*’ = invariant (49%); ‘:’ = highly similar (16%); ‘.’ = similar (10%). Alignment of the results predicting helical motifs (indicated as ‘*’) from a series of secondary structure prediction programmes (B). The green and red bars indicate that the sequence taken for simulation of TMD1 and TMD2, respectively, has a helix. The black line indicates the amino acids of the loop-region, the blue bars indicate the extramembrane parts.