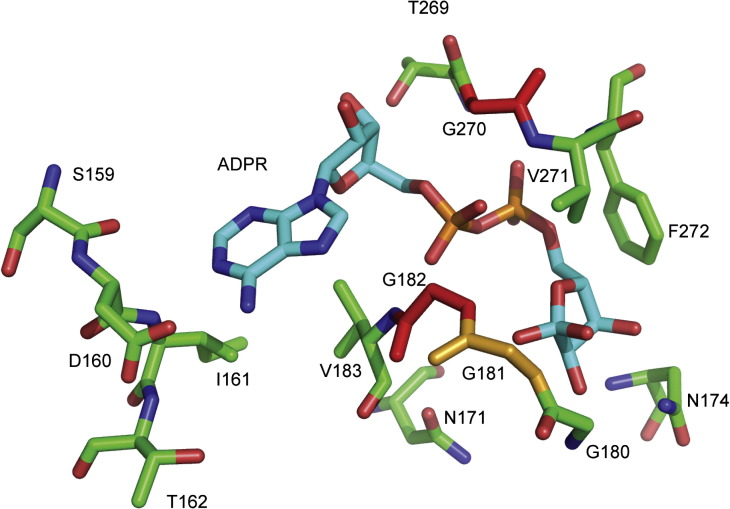
Fig. 4.
Structure prediction of the MDO1 ligand binding site. The ADP-ribose bound to the MDO1 ligand-binding pocket is shown as predicted, using SARS-CoV macro domain as model. Sites of mutations inactivating ADP-ribose binding (residues G182 and G270) are highlighted in red, and the site of mutation reducing binding (G181) is shown in orange. In the SFV macro sequence, positions 31 and 32 correspond to MDO1 residues 181–182, and position 112 corresponds to MDO1 residue 270. The residues corresponding to the additionally mutated sites in SFV are also seen in this rendering: residues D10, N21, and N24 in the SFV sequence correspond to MDO1 D160, N171, and N174, respectively.