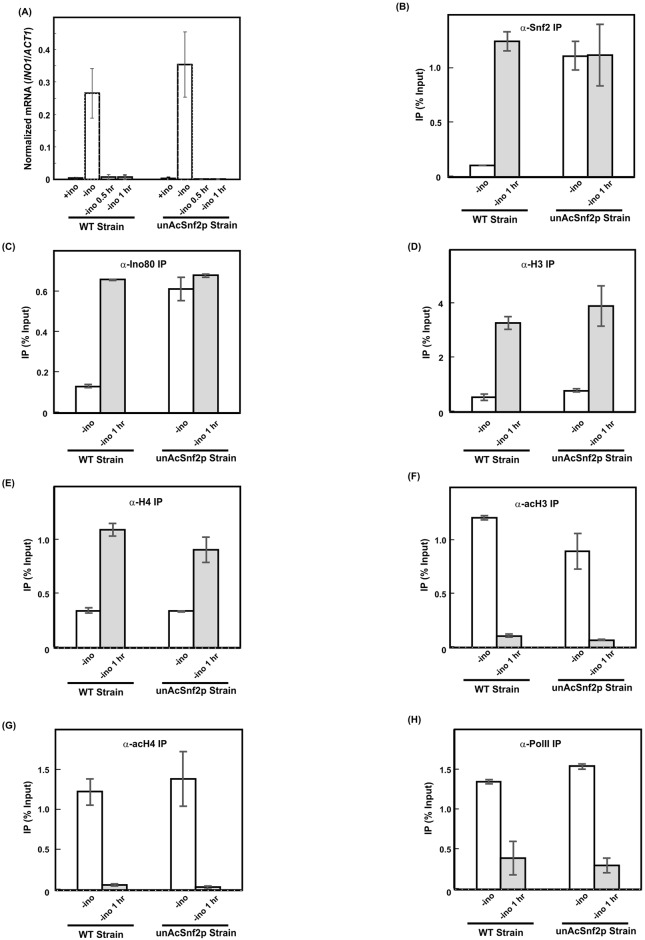
Fig 4. The INO1 induction and chromatin remodeling activities are indepedent of Snf2p acetylation when switching from inducing conditions to repressing conditions.
(A) INO1 mRNA was examined by qRT-PCR analysis. Cells were grown in SC media to mid-log phase (A600 = 1.0) and were subsequently washed and resuspended in inducing conditions for 30 min or 1hr, respectively. Subsequently, cells were switched to repressing conditions for another 1 hr 30 min or 1 hr prior to collection. The expression ratio for INO1 mRNA/ACT11 mRNA is graphed as mean standard ± deviation. All experiments were repeated at least three times, and in each experiment PCR reactions were done in triplicate. (B-H) Real-time PCR analysis of DNA immunoprecipitated through Chromatin Immunoprecipitation (ChIP) with an antibody against (B) FLAG-tagged Snf2p (α-Snf2p), (C) Ino80p (α-Ino80p), (D) Histone H3 (α-H3); (E) Histone H4 (α-H4); (F) Acetylated Histone H3 (α-acH3); (G) Acetylated Histone H4 (α-acH4); (H) RNA polymerase II (α-Pol II) at the INO1 promoter for WT cells and unAcSnf2p cells that were grown to mid-log phase (A600 = 1.0) in SC and were subsequently washed and subjected to inducing (-ino; open bars) or switching (1 hr inducing followed by 1 hr repressing conditions) (-ino 1hr; light grey bars) conditions, respectively, for total 2 hours incubation prior to collection. All experiments were performed with three repeats and all PCR reactions were done in at least twice. The IP for the INO1 promoter is graphed as mean ± standard deviation normalized to input and mock.