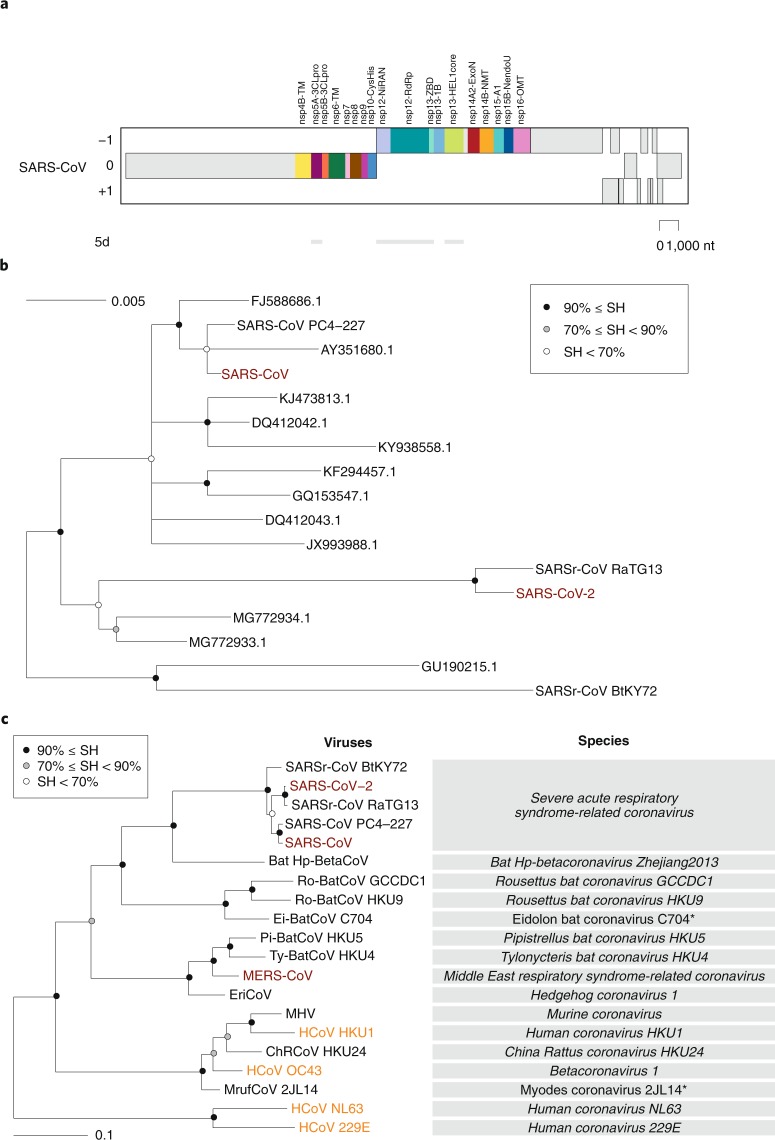
Fig. 2. Phylogeny of coronaviruses.
a, Concatenated multiple sequence alignments (MSAs) of the protein domain combination44 used for phylogenetic and DEmARC analyses of the family Coronaviridae. Shown are the locations of the replicative domains conserved in the ordert Nidovirales in relation to several other ORF1a/b-encoded domains and other major ORFs in the SARS-CoV genome. 5d, 5 domains: nsp5A-3CLpro, two beta-barrel domains of the 3C-like protease; nsp12-NiRAN, nidovirus RdRp-associated nucleotidyltransferase; nsp12-RdRp, RNA-dependent RNA polymerase; nsp13-HEL1 core, superfamily 1 helicase with upstream Zn-binding domain (nsp13-ZBD); nt, nucleotide. b, The maximum-likelihood tree of SARS-CoV was reconstructed by IQ‑TREE v.1.6.1 (ref. 45) using 83 sequences with the best fitting evolutionary model. Subsequently, the tree was purged from the most similar sequences and midpoint-rooted. Branch support was estimated using the Shimodaira–Hasegawa (SH)-like approximate likelihood ratio test with 1,000 replicates. GenBank IDs for all viruses except four are shown; SARS-CoV, AY274119.3; SARS-CoV-2, MN908947.3; SARSr-CoV_BtKY72, KY352407.1; SARS-CoV_PC4-227, AY613950.1. c, Shown is an IQ‑TREE maximum-likelihood tree of single virus representatives of thirteen species and five representatives of the species Severe acute respiratory syndrome-related coronavirus of the genus Betacoronavirus. The tree is rooted with HCoV-NL63 and HCoV-229E, representing two species of the genus Alphacoronavirus. Purple text highlights zoonotic viruses with varying pathogenicity in humans; orange text highlights common respiratory viruses that circulate in humans. Asterisks indicate two coronavirus species whose demarcations and names are pending approval from the ICTV and, thus, these names are not italicized.