Fig. 1. Human light chains, S35 and S38.
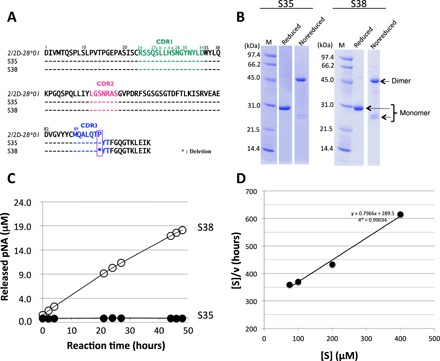
(A) Comparison of germline (2/2D-28*01), S35, and S38. The sequence of the Vκ region (amino acids 1 to 95) of S35 having Pro95 is identical to the germline (2/2D-28*01). On the other hand, S38 lacks the Pro95. (B) SDS-PAGE analysis with CBB (coomassie brilliant blue) staining. Bands of about 46 and 26 kDa correspond to the dimer and monomer, respectively, under the nonreduced condition. Only a 30-kDa band under the reduced condition corresponds to the monomer. Bands other than the monomer of the light chains were hardly observed under the reduced condition. (C) Time course of the cleavage reaction. The substrate: R-pNA (200 μM). Light chain: S38 (10 μM: open circle) and S35 (10 μM: closed circle). Out of five substrates (R-, E-, L-, A-, and FL-pNA), only R-pNA was cleaved by S38. The S35 did not cleave any substrates at all. (D) Kinetic analysis by S38 light chain. The concentration of the S38 light chain was fixed at 5 μM, and that of the R-pNA substrate varied from 75 to 400 μM at 37°C. [S] indicated concentration of R-pNA; [V], initial rate. The Hanes-Woolf plot demonstrates that the cleavage reaction by the S38 light chain fits the Michaelis-Menten kinetics equation, indicating that the reactions are enzymatic.
