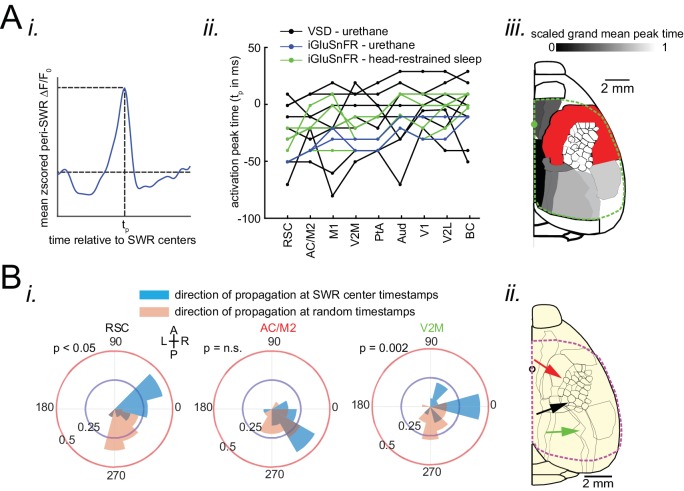
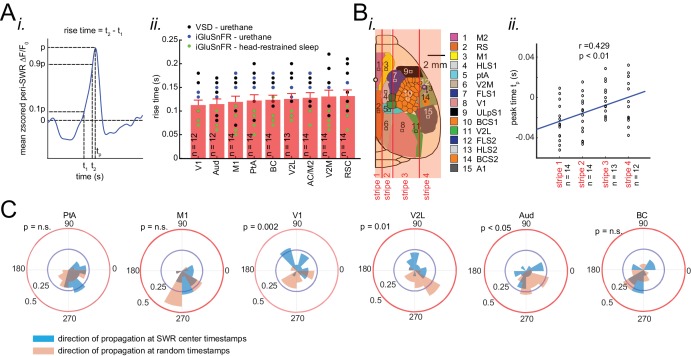
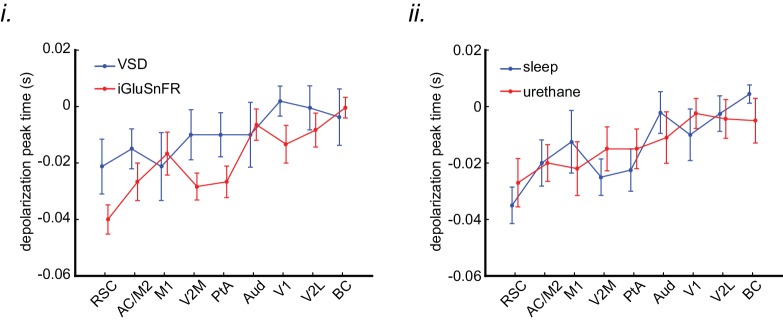
Figure 4. Neocortex tends to sequentially activate from medial to more lateral regions around SWRs.
(A) (i) Demonstration of how peri-SWR neocortical activation peak time (tp) was quantified. The mean peri-SWR neocortical activity trace was generated for each region (blue trace) and the timestamp of the peak was defined as tp. (ii) Peri-SWR activation peak timestamp (tp) relative to SWR centers (0s-time) across neocortical regions sorted in ascending order. Each line graph represents one animal. tp values were not detected in some regions and in some animals (three data points in total), mainly because there was not a strong activation in those regions. Such missing data points were filled by average of available data points in the same region and in other animals (repeated measure ANOVA: F8,104 = 8.357195, p<0.0001; post-hoc test for linear trend: slope = 0.003252024 s/region, p<0.0001). (iii) Spatial map of peri-SWR activation peak time across all animals (n = 14) indicating a medial-to-lateral direction of activation. The red area was not included in this analysis because it was not activated strong enough to yield a reliable result. (B) (i) Circular distributions represent the direction of propagating waves of activity in three distinct neocortical regions at hippocampal SWR (blue distribution) and at random timestamps generated by shuffling inter-SWR time intervals (red distribution). 180–0 and 90–270 degrees represent the medio-lateral and antero-posterior axes, respectively. P-values come from Kuiper two-sample test. (ii) Schematic of propagation directions measured at SWR timestamps in three neocortical regions located in the medial neocortex. This figure has two figure supplement.