FIGURE 2.
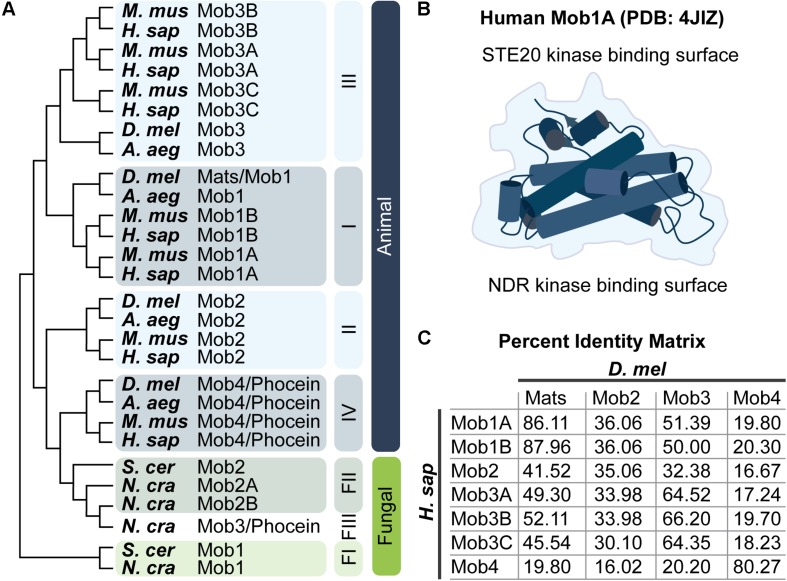
Mob family proteins are conserved from yeast to humans. (A) Molecular phylogenetic analysis of Mob protein sequences from the indicated species. Tree was generated using MEGA X (Jones et al., 1992; Kumar et al., 2018) as described by Hall (2013). Animal Mob proteins cluster into four distinct classes (I–IV). The relative divergence between these classes is not revealed in this limited comparison of Mobs from four animal and three fungal species – three fungal Mob classes are indicated (FI–FIII). For a thorough discussion of the evolutionary history of Mob family proteins consult the work of Vitulo et al. (2007) and Ye et al. (2009). A. aeg, Aedes aegypti; D. mel, Drosophila melanogaster; H. sap, Homo sapiens; M. mus, Mus musculus; N. cra, Neurospora crassa; S. cer, Saccharomyces cerevisiae. Refer to the Pfam database which maintains a manually curated annotation of the Mob family proteins (PF03637; El-Gebali et al., 2019). Mob family protein sequences are readily available at uniprot.org by searching for “Mob kinase activator” (The UniProt Consortium, 2019). (B) Artistic sketch of the human Mob1A protein (PDB: 4JIZ) reported by Rock et al. (2013). Mob proteins adopt a conserved globular fold with a core that consists of a four alpha-helix bundle, we refer to this fold as the “Mob family fold.” Binding to NDR kinases or STE20 kinases takes place on distinct Mob surfaces as indicated. (C) Percent identity matrix between human and fly Mob proteins.