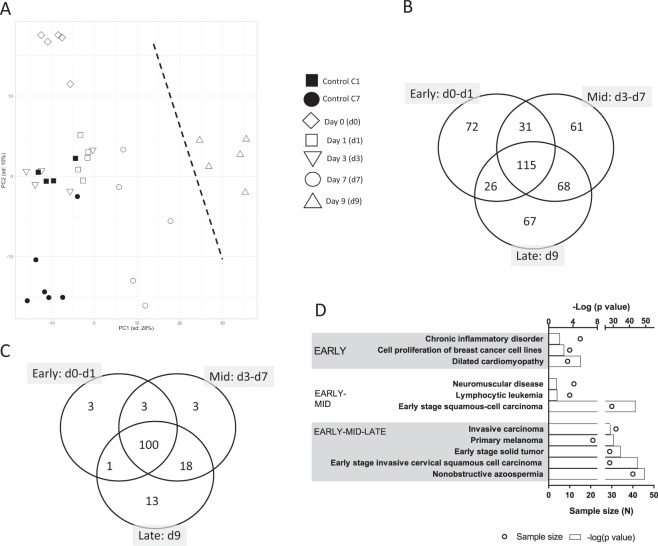
Figure 2.
The whole genome miRNA profile responding to lethal radiation. (A) Principal component analysis (PCA) of the whole genome miRNA data shows a clear separation between d9 post-TBI and rest of the time points including the controls. A dotted line marks the separation. (B) Venn diagram shows the longitudinal distribution of differentially expressed miRNAs across three time phases post-TBI, namely, Early (d0–d1), Mid (d3–d7) and Late (d9). The largest number of differentially expressed miRNAs (115 miRNAs) are found at the cross section of the three phase. Nevertheless, a considerable number of miRNAs are exclusively regulated during each of the three phases. For instance, there are 72, 61 and 67 miRNAs exclusively regulated in the Early, Mid and Late phases, respectively. (C) Venn diagram shows the longitudinal distribution of significantly enriched networks across three time phases post-TBI, namely, Early (d0–d1), Mid (d3–d7) and Late (d9). Nearly all of these networks are concentrated at the cross section of all three time phases. (D) Top 5 most significantly enriched networks during the Early, Early-Mid and Early-Mid-Late phases are shown in this bar chart. Early and Early-Mid phases enlist only 3 networks, so all of them are included here. Top horizontal axis represents the −log(p value), and the bar charts represents the hypergeometric p-values converted to log10 associated with individual networks. The bottom horizontal axis represents the sample size (N), and the open circles represent the number of miRNAs associated with individual networks.