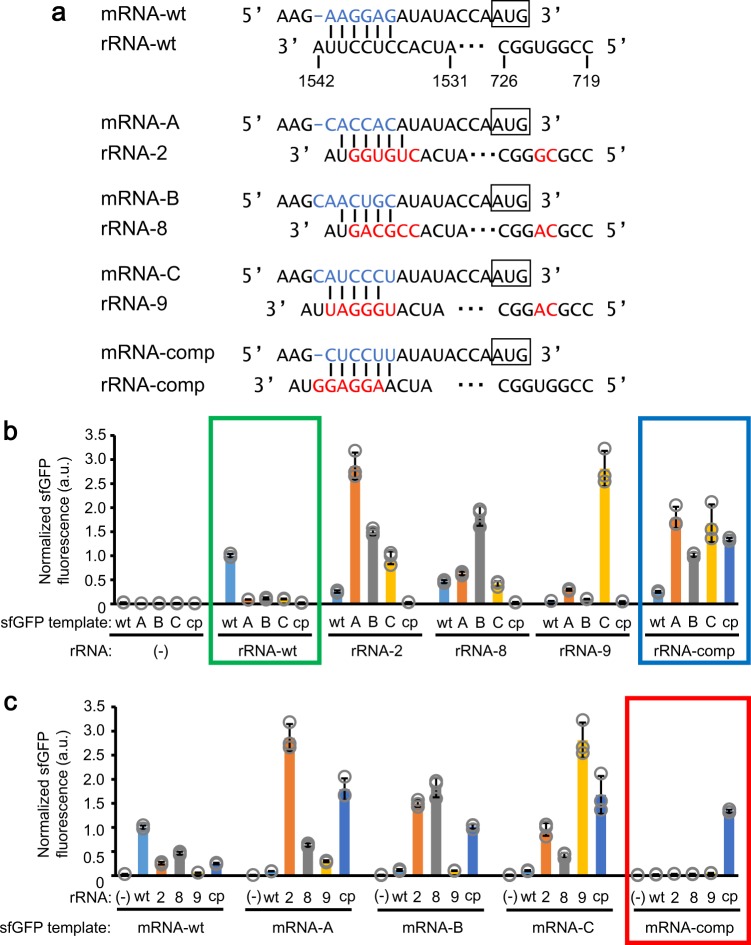
Fig. 3. A mutational study of 16S rRNA to search for applicable orthogonal SD/anti-SD pair for differentiating “host” and “newly assembled” ribosomes.
a Sequences of 16S rRNA and mRNA examined in this study. SD sequences are indicated by light blue characters, and mutated nucleotides in 16S rRNA by red characters. b, c Synthesis of sfGFP using various SD/anti-SD pairs. Increase in sfGFP fluorescence after 4 h incubation was normalized by dividing by the average value of the control reaction. Graphical display is different, but the same data were used in (b) and (c). Symbols “wt”, “A”, “B”, “C”, and “cp” for sfGFP template represent mRNA-wt, mRNA-A, mRNA-B, mRNA-C, and mRNA-comp, respectively, as shown in (a). Symbols “wt”, “2”, “8”, “9”, and “cp” for rRNA represent rRNA-wt, rRNA-2, rRNA-8, rRNA-9, and rRNA-cp, respectively, as shown in (a). A green box indicates a data set obtained using rRNA-wt, a blue box indicates a data set obtained using rRNA-comp, and a red box indicates a data set obtained using mRNA-comp. Error bars indicate standard deviation of triplicate measurements. Each dot represents individual observed value.