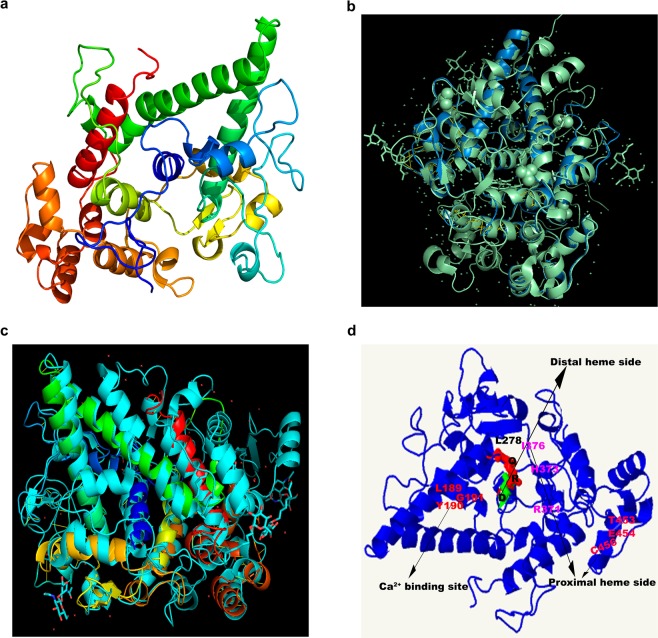
Figure 1.
In silico reconstruction of the TrotCOX protein structure compared with the known reference proteins for the peroxidase-cyclooxygenase superfamily. (a) TrotCOX structure based on the 2.75 Å resolved x-ray diffraction of BtLPO. (b) Comparison of the in silico structure of the TrotCOX protein and the 2.75 Å resolved x-ray diffraction of BtLPO showing the perfect overlap of the alpha-helices embedding the heme sides. In blue the TrotCOX, in light green the BtLPO. (c) Comparison of the in silico structure of the TrotCOX protein and the 2.75 Å resolved x-ray diffraction of OaCOX1 showing the perfect overlap of the alpha-helices embedding the heme sides. Rainbow colored helices correspond to TrotCOX, the light-blue colored helices correspond to OaCOX1. (d) Illustration of the conserved catalytic sites described in the text. Q, R, D and L278 black letters refer to the conserved amino acids of the two distal heme sides with L278 being the mutated original E. Pink and light red letters indicate the conserved amino acids in the two proximal heme sides. The red letters indicate the amino acids forming the calcium binding site.