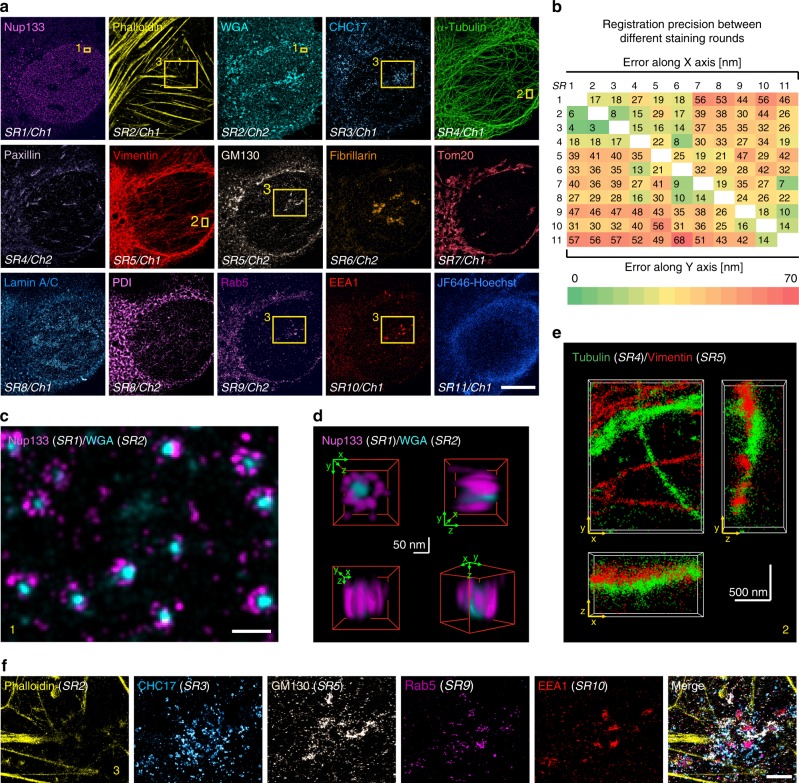
Fig. 2. Multi-target 3D dSTORM of a single U2OS cell.
a A U2OS cell with 15 targets labeled and acquired by 11 staining rounds (SR). Two channels (Ch) can be detected per SR, but not always each channel can be used for final analysis. In addition, to get optimal signal for some sensitive labels (e.g., GFP nanobodies against the Ypet-tag of Nup133), it was beneficial to not stain targets in the second channel (see Supplementary Note 2). Boxes denote regions magnified in c, e, and f. Another three experiments were carried out with similar designs and outcomes. Further details see Methods section. b Heatmap representing the registration error along the x (upper matrix part) and the y axis (lower matrix part) reveals precise registration, in particular, between images of adjacent staining rounds (arranged along the white diagonal fields). c Magnified view of boxed region 1 in a with merged peripheral Nup133 and central WGA nuclear pore labeling demonstrating that super-resolution images from consecutive SRs can be registered with high precision. d 3D views of a nuclear pore complex (from the multiplex experiment in a) demonstrate the high lateral and axial resolution and registration precision. e Merged tubulin and vimentin 3D images from consecutive SRs, demonstrating that super-resolution images can be acquired and registered in 3D (boxed region 2 in a). f Five-color merge of boxed region 3 in a: phalloidin-labeled actin filaments, CHC17-labeled clathrin-coated structures, GM130-labeled proximal Golgi apparatus, endosome-associated proteins Rab5 and EEA1. Scale bars correspond to 10 µm in a, 200 nm in c, 50 nm in d, 500 nm in e, and 2 µm in f.