Abstract
Melioidosis is a severe disease caused by the environmental bacterium Burkholderia pseudomallei that affects both humans and animals throughout northern Australia, Southeast Asia and increasingly globally. While there is a considerable degree of genetic diversity amongst isolates, B. pseudomallei has a robust global biogeographic structure and genetic populations are spatially clustered in the environment. We examined the distribution and local spread of B. pseudomallei in Darwin, Northern Territory, Australia, which has the highest recorded urban incidence of melioidosis globally. We sampled soil and land runoff throughout the city centre and performed whole-genome sequencing (WGS) on B. pseudomallei isolates. By combining phylogenetic analyses, Bayesian clustering and spatial hot spot analysis our results demonstrate that some sequence types (STs) are widespread in the urban Darwin environment, while others are highly spatially clustered over a small geographic scale. This clustering matches the spatial distribution of clinical cases for one ST. Results also demonstrate a greater overall isolate diversity recovered from drains compared to park soils, further supporting the role drains may play in dispersal of B. pseudomallei STs in the environment. Collectively, knowledge gained from this study will allow for better understanding of B. pseudomallei phylogeography and melioidosis source attribution, particularly on a local level.
Subject terms: Microbial genetics, Bacterial genetics, Infectious-disease epidemiology
Introduction
Burkholderia pseudomallei is an environmental Gram-negative bacillus and the causative agent of melioidosis, a potentially fatal infection of humans and animals1. Regions of high B. pseudomallei endemicity predominantly include Southeast Asia and northern Australia, though the bacterium is also increasingly found in other tropical regions including the Indian subcontinent, Africa and the Americas2. Melioidosis is typically considered noncommunicable with direct person-to-person transmission and zoonotic disease being remarkably rare3,4. Nearly all B. pseudomallei infections are caused by a single direct exposure event to contaminated soil or surface water in the environment and individual cases of melioidosis are typically the result of infection by different strains of the bacterium5.
A limited geographic dispersal of B. pseudomallei strains has also been identified and the bacterium is now recognised as being ecologically established and spatially clustered in the environment6–8. This is in spite of the frequent opportunities B. pseudomallei has to spread within the water table, via agricultural and migratory animals, or in transported soil6,9–11. This restricted geographical distribution has resulted in distinct genetic populations of the bacterium, which are evident despite high levels of gene recombination and sequence type (ST) diversity9,12,13. While the high rate of genetic diversity and recombination previously hindered the examination of B. pseudomallei populations using traditional typing methods the development of whole-genome sequencing (WGS) has facilitated the examination of genetic populations of the bacterium on a fine-scale8,12,14,15. This has allowed for population and evolutionary inferences to be made on both a global and local level. WGS, in conjunction with multi-locus sequence typing (MLST) data, has revealed distinct geographical partitioning between Australian and Southeast Asian isolates separated by Wallace’s Line12,13,16. Additionally, no shared environmental STs have been identified within the Northern Territory and adjacent Queensland, northern Australia, with distinct B. pseudomallei population structures identified in the two regions using Bayesian MLST-based analysis9.
While more than 450 MLST types have now been classified in the Northern Territory, Australia, (https://pubmlst.org/bpseudomallei/) five ST’s have been shown to comprise 90% of the overall sequence type abundance in the Darwin region of the Northern Territory, Australia and the maximum geographic distance identified between environmental isolates of the same strain is typically in the range of 50 linear kilometres6,7,17. This again supports the idea that despite the high degree of diversity, populations of B. pseudomallei are ecologically established and not widely dispersed in the environment. Despite the high degree of ST diversity in the Northern Territory environment a distinct genetic population structure of the bacterium has yet to be identified there.
Within the “Top End” of the Northern Territory, Australia, B. pseudomallei is frequently isolated in the environment and there have been more than 1,150 culture-confirmed human cases of melioidosis diagnosed since 19895,6,18. The coastal capital city of Darwin (12°S latitude) has by far the highest incidence of melioidosis reported for any urban environment globally, with rates up to 50 per 100,000 annually19. Apart from soil, B. pseudomallei is also frequently detected in water. A third of tested unchlorinated rural water bores, of which there are over 5,000 in the Darwin surrounds, have been shown to be B. pseudomallei positive20 and these have been implicated in clusters of melioidosis infections in the past4,21,22. Moreover, studies into seasonal disease correlates in northern Australia have demonstrated an association between the frequency of cases and the nature and timing of rainfall-related events23. Contaminated water and B. pseudomallei’s movement within the water table have recently been demonstrated as a potentially significant environmental reservoir and distribution tool for the bacterium there11,24. Environmental isolates recovered from groundwater seeps in Townsville, Queensland, Australia were later linked to clinical isolates using molecular typing11,24.
Since storm-water is known to capture and leach what is in the land, including particles, contaminants and bacteria, it may provide a more appropriate indication of catchment and B. pseudomallei distribution, as the bacterium is able to further disperse along drainage lines25–27. As a result of B. pseudomallei’s heterogeneous distribution in the environment, the identification of new areas endemic for melioidosis may be more effectively determined by analysing surface runoff and storm-water than by the analysis of random soil samples11,27,28.
The aim of this study was to examine the distribution and local population structure of B. pseudomallei in urban Darwin, Northern Territory, Australia targeting public parks and drains. We sampled soil and land runoff throughout the city and did WGS analysis on cultured isolates. By combining genome variant analysis and Bayesian clustering methods we examined the local phylogenetic structure of B. pseudomallei and used spatial hot spot analyses to examine the spatial clustering of the bacterium on a small geographic scale. We hypothesised that B. pseudomallei would be well-established throughout the urban Darwin environment and there would be a considerable amount of ST diversity amongst isolates, particularly at drain catchment sites. However, we predicted that genetic spatial clustering of B. pseudomallei would not occur on such a localised geographic scale.
Results
Presence of B. pseudomallei in urban Darwin drains
From the 42 drain sites surveyed in the first sampling round in the early wet season, 2016, 19 (19/42, 45.2%) had water available for collection. Five of these 19 sites only contained enough water to collect one replicate. Of the 47 total water samples collected, five samples (5/47, 10.6%) were qPCR and/or culture positive at three sites (3/19, 15.8%). Two of the samples (2/47, 4.3%) were qPCR positive only at two sites (2/19, 10.5%).
In contrast, water was available for collection in all but six drains (36/42, 85.7%) in the second sampling round at the end of the wet season (Feb-Mar 2017). From the 108 total water samples collected 40.7% were culture and/or qPCR-positive, with 55.6% of drain sites being positive. Of these, 19.4% of samples were qPCR positive only at 38.9% sites (Table 1). From the 210 total soil samples collected from areas surrounding drains during the wet season, 30.5% were culture and/or qPCR-positive from 66.7% of sites. (Table 1, Fig. 1a).
Table 1.
B. pseudomallei positive sites and samples at the end of the rainy season based on culture and qPCR detection methods.
| Drain soil | Park soil | Total soil (park and drain) | Total water (drains) | ||
|---|---|---|---|---|---|
| Samples | Culture positive only | 5.2% (11/210) | 1.5% (7/450) | 2.7% (18/660) | 0% (0/108) |
| qPCR positive only | 5.7% (12/210) | 6.4% (29/450) | 6.2% (41/660) | 19.4% (21/108) | |
| Total positive by culture and/or qPCR | 30.5% (64/210) | 39.6% (178/450) | 36.7% (242/660) | 40.7% (44/108) | |
| Sites | Total positive by culture and/or qPCR | 66.7% (28/42) | 82.2% (37/45) | 74.7% (65/87) | 55.6% (20/36) |
Figure 1.
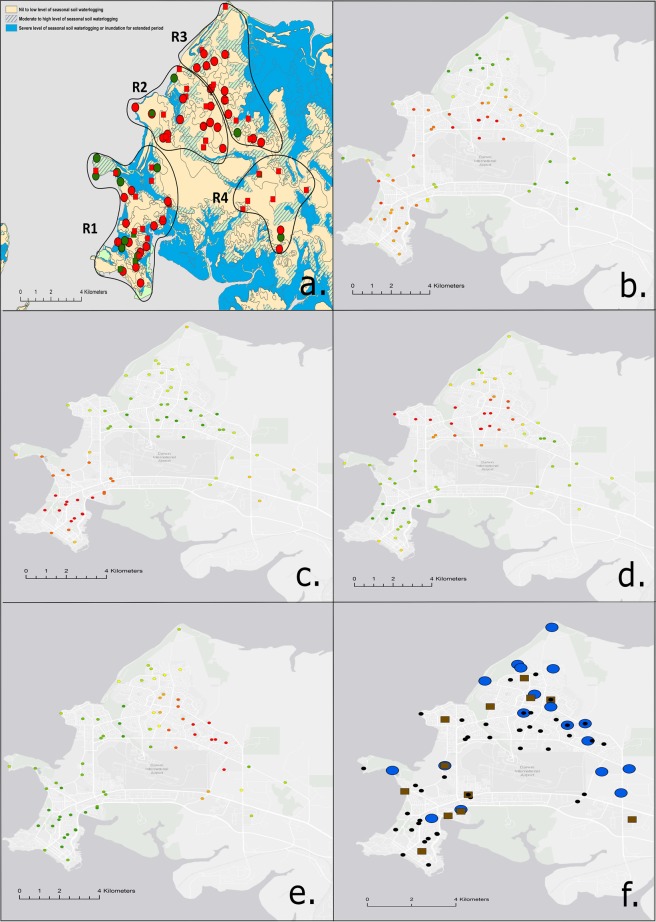
The four urban Darwin regions specified by catchment areas and direction of surface runoff at each survey site (a). Levels of seasonal surface runoff and areas of waterlogging are indicated by colour (yellow; nil to low levels of seasonal waterlogging, green stripes; moderate to high levels, blue; severe levels of waterlogging to fully inundated over extended period). B. pseudomallei positive and negative park sites (n = 45, red and green circles, respectively) and positive and negative drain sites (n = 42, red and green squares, respectively) surveyed during the 2017 rainy season (Feb-March). Getis-Ord Gi* Hot Spot Analysis (b–e). Areas with significant “hot” spots are indicated by dark red circles while “cold” spots are shown in green ((b) ST-36, (c) ST-109, (d) ST-327, (e) ST-553). Significant association of Bayes Cluster 1 with drain sites (blue circles – water and soil) as opposed to parks (brown squares - soil) (f). Bayes cluster 1 negative sites are denoted by black circles (ArcGIS v.10.4.1 (ESRI)).
Incidence of B. pseudomallei in urban Darwin parks
From the 450 soil samples collected throughout urban parklands during the end of the rainy season, 39.6% of samples were culture and/or qPCR-positive, with 37/45 (82.2%) of surveyed sites being positive overall (Table 1, Fig. 1a).
Overall, significantly more water samples were B. pseudomallei positive only by qPCR compared to soil (parks and drains) (19.4% compared to 6.2%, Fisher’s exact test P < 0.001).
Sequence type diversity
From the 135 B. pseudomallei environmental soil and water isolates selected for WGS we identified 35 distinct MLST genotypes (Supplementary Table S1). ST-36 was the most frequently observed molecular type (n = 42), followed by ST-109 (n = 22), ST-327 (n = 11) and ST-553 (n = 8). Eight of the ST types were novel, seven of which were identified in isolates collected from soil. Of the STs previously identified, all but one strain (ST-362) had also been isolated from a Darwin Prospective Melioidosis Study (DPMS) clinical patient5.
There were 24 STs that were identified in soil only, while four STs were isolated in water but not soil (ST-131, ST-456, ST-472, ST-1654). Only seven strains were isolated from both sample types, which included the four most frequently isolated STs (ST-36, ST-109, ST-144, ST-320, ST-327, ST-553, ST-616). (Supplementary Table S1). More STs were identified overall in soil than were in water (82.9% compared to 31.4% of the 35 STs recovered) (Table 2). Accounting for isolates recovered per category, drain soil had a higher overall ST diversity compared to park soil (51.2% compared to 30.1%, Fisher’s Exact test P = 0.029).
Table 2.
Total number of samples and STs identified amongst the 135 B. pseudomallei study isolates based on site (42 drain and 45 park sites) and sample types.
| Drain soil | Park soil | Total soil (park and drain) | Drain water | Total drains (water and soil) | ||
|---|---|---|---|---|---|---|
| Percentage isolates (%) | Vs. total isolates examined (n = 135) | 30.4% (41/135) | 54.1% (73/135) | 84.4% (114/135) | 15.6% (21/135) | 45.9% (62/135) |
| Percentage STs (%) identified | Vs. total STs (n = 35) | 60.0% (21/35) | 62.9% (22/35) | 82.9% (29/35) | 31.4% (11/35) | 68.6% (24/35) |
| Vs. number of isolates from that category | 51.2% (21/41) | 30.1% (22/73) | 25.4% (29/114) | 52.4% (11/21) | 38.7% (24/62) |
Population structure of B. pseudomallei across urban Darwin
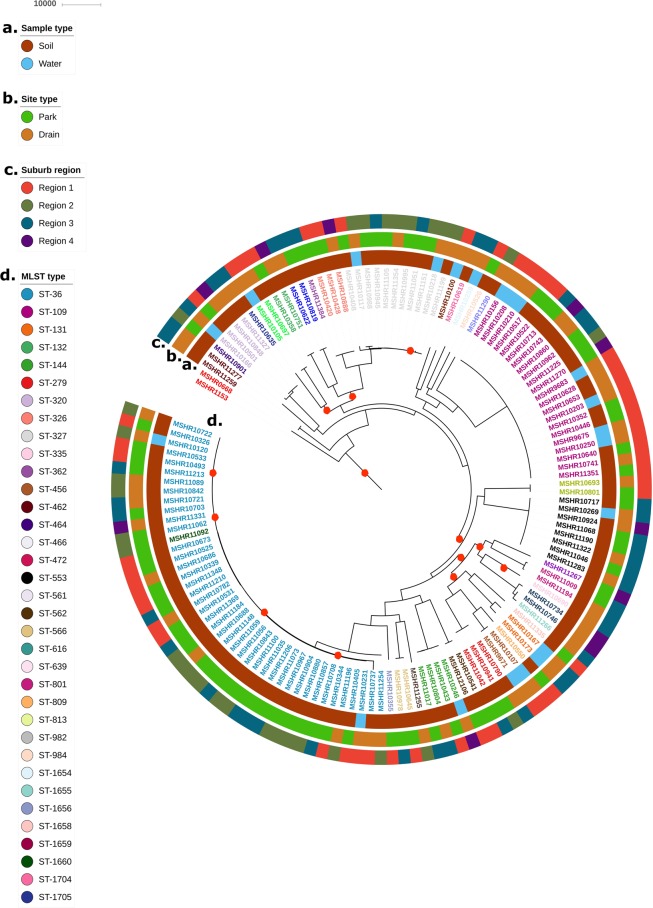
Despite the high levels of recombination in the B. pseudomallei genome, maximum parsimony (MP) phylogenetic reconstruction of the 134,032 orthologous biallelic single-nucleotide polymorphisms (SNPs) and insertion and deletion events (InDels) identified robust clustering of sequence types (Fig. 2). This was true for all but one isolate, MSHR11092, which clustered with the ST-36 group despite being designated as a novel strain type, ST-1660 (Fig. 2). However, compared to the ST-36 isolates, MSHR11092 contained a single-locus variant at the nitrite extrusion protein (narK) MLST gene loci (https://pubmlst.org/bpseudomallei/). Grouping of isolates was generally consistent with the reconstructed maximum likelihood tree29 (Supplementary Fig. S1).
Figure 2.
Maximum parsimony phylogeny of 137 B. pseudomallei genomes. MP reconstruction of 134,032 core-genome orthologous SNPs and InDels belonging to environmental survey isolates. MSHR1153 was used as the reference strain and the tree was rooted at MSHR0668. The sample type (a), site type (b), suburb region (c) and MLST type (d) of each isolate are labelled as described. Red circles on branches denote bootstrap values <80.
While isolates clustered by ST, they did not group by whether they were isolated from soil or water, nor by whether they were collected from a drain or park site. There was also no observable clustering by region based on phylogenetic reconstruction of isolates.
The population structure of the 135 isolates was further defined using RhierBAPS based on a core genome SNP mapping alignment, which divided the 135 isolates into five primary sequence clusters (BAPS hierarchical level 1). These were further subdivided into 16 lineages (BAPS level 2) (Supplementary Fig S2). Three of the primary clusters at level 1, clusters 1, 2 and 3, were all comprised of multiple different STs, while ST-109 and ST-553 were uniquely assigned as cluster 4 and 5, respectively. Bayes cluster 1 contained the largest percentage of STs of any cluster (68.6%, 24/35). The five clusters were largely congruent with STs and the reconstructed phylogeny. However, clusters 1 and 2 included several STs that were distributed across different branches of the MP tree.
B. pseudomallei is spatially clustered within the urban Darwin environment
There was strong evidence of spatial clustering in the environment for three of the four common urban Darwin STs for which we performed hot spot analysis, including ST-109, ST-327, and ST-553 (Getis-Ord Gi* statistic (GiZ) score >1.96, p < 0.01) (Fig. 1b–e). No significant hot spots were identified for ST-36 isolates, which were well-dispersed across sample sites (Fig. 1b). Locations and sizes of hot spots varied amongst the three STs that clustered in the environment. For ST-109, we detected a large hot spot in the city centre in the southwest region of urban Darwin (Region 1, (Fig. 1c)), while significant hot spots were observed for ST-327 (Fig. 1d) and ST-553 (Fig. 1e) in the western and eastern regions of the Darwin northern suburbs (Region 2 and Region 3, respectively). This correlated with logistic regression models of ST-109 and ST-327, which also indicated that ST-109 and ST-327 were strongly associated with Regions 1 and 2, respectively (ST-109; OR 5.7; 95% CI 1.28–25.42; p value 0.022 and ST-327; OR 7.1; 95% CI 1.53–33.30; p value 0.012). ST-553 was only isolated from the eastern northern suburb area (Region 3) (Fisher’s Exact test, p value 0.002). No hot spots were identified for any of the common STs we examined in the less densely populated Region 4 and there was strong evidence for decreased ST diversity by site in this region when compared to Regions 1 and 2 (Negative binomial regression, p value 0.024 and p value 0.006, respectively).
Hot spot analysis was also performed on the five primary RhierBAPS clusters. No significant clustering was identified for clusters 1 or 2. Likewise, since clusters 4 and 5 each contained only a single unique ST, the hot spots identified for these clusters were identical to their corresponding MLST types (ST-109- Fig. 1c, ST-553- Fig. 1e). While we observed no spatial clustering of ST-36 isolates individually (Fig. 1b) RhierBAPS cluster 3, which was comprised of ST-36, ST-566, ST-1656, ST-1659, and ST-1660, showed a significant hot spot in the central northern suburb region (Supplementary Fig. S3).
Association of sequence types and genetic diversity with sample and site variables
B. pseudomallei ST-36 was strongly associated with soil (OR 5.7; 95% CI 1.82–17.8; p value 0.003) (intra-cluster correlation ICC = 0.13). This correlated with Bayes primary cluster 3 (ST-36, ST-566, ST-1656, ST-1659, and ST-1660), which was also strongly associated with soil (OR 2.6; 95% CI 1.12–5.89; p value 0.026) (ICC = 0.17). Additionally, occurrence of ST-109 was positively associated with water (OR 2.51, 95% CI; 1.0–6.32, p; 0.05). The odds to find an isolate belonging to Bayes primary cluster 1 which contained the largest ST diversity was 2.7 times larger at drain sites compared to parks (OR 2.7; 95% CI 1.23–5.94; p value 0.13) (ICC = 0.09) (Fig. 1f).
Moreover, the odds to recover more than one ST at a site was 3 times higher in parks compared to drains (OR 3.1; 95% CI 1.11–8.43; p value 0.030). Consistent with this, there was an increased probability of finding more than one ST at sites immediately adjacent to a sport’s field (OR 4.4; 95% CI 1.17–16.15; p value 0.03).
Association of environmental ST clusters with clinical patient isolates
Lastly, the odds of having a ST-553 clinical case with patient residence in Region 3 were 4.2 times higher compared to the other Darwin regions (OR 4.2; 95% CI 2.23–7.9; p value <0.001). This matched the Region 3 cluster of ST-553 environmental survey isolates. Regional environmental clusters identified for ST-109 (Region 1) and ST-327 (Region 2) were not associated with the suspected location of infection for urban Darwin ST-109 and ST-327 clinical patient isolates.
Discussion
In this study we analysed the distribution and local population structure of B. pseudomallei in urban Darwin, Northern Territory, Australia by sampling public parks and drains. B. pseudomallei was detected from more than 80% of park and drain sites sampled during the wet season, indicating it is ubiquitous in the urban Darwin environment and that targeting drain sites will be useful for future environmental monitoring of the bacterium. While WGS data demonstrated a large amount of genetic diversity amongst survey isolates, our results indicate that certain STs are more widespread in the urban Darwin environment. Additionally, we showed that WGS and Bayesian analysis of SNP data can be used to examine the phylogeography of B. pseudomallei isolates on a small geographic scale, with results suggesting there are distinct genetic populations of B. pseudomallei within urban Darwin and that spatial clustering of populations exists over a remarkably restricted geographical area.
While tropical soils are the known natural reservoir for B. pseudomallei, during periods of heavy rainfall and increased surface discharge the bacterium can be leached out of the soil and channelled into drainage areas along with other eroded particulate matter25,26,30. Contaminated water and movement of B. pseudomallei within the water table have now been established as a significant environmental source and potential distribution tool for the bacterium in melioidosis endemic regions, including southern Laos and northern Queensland, Australia26–28. Accordingly, our results indicate that surface runoff is a potential source of melioidosis infection within urban Darwin and may play a significant role in dispersal of B. pseudomallei there, both along drainage lines and via the moving water table25,31. Land use is known to play an integral role in the transfer of bacteria through soils to downstream aquatic systems and catchment areas26,30,32. As urban Darwin continues to develop and expand, increased construction may ultimately lead to additional soil erosion and runoff. This could potentially affect the distribution and dispersal of the bacterium there, particularly during periods of heavy rainfall26,31. Thus, the potential for increased rates of B. pseudomallei transmission and its propagation to uncontaminated areas should be considered as the city continues to grow. Similar considerations apply to future urban development throughout tropical northern Australia.
Previous phylogenetic investigations of B. pseudomallei have shown that Australian strains are ancestral to those found in Southeast Asia and globally, with the highest degree of genetic diversity identified in the Northern Territory of Australia6,9,12,13. Nevertheless, the nature and mechanisms for the global spread of B. pseudomallei remain to be elucidated, with explanations for autochthonous cases in the USA still proving elusive33. We identified 35 distinct B. pseudomallei STs as part of the investigation, further demonstrating that the bacterium is genetically diverse and well-established in the Darwin environment. Despite this, ST-36 and ST-109 comprised nearly 50% of the 135 isolates examined and ST-327 and ST-553 were the only additional molecular types for which we identified more than five isolates. These results are consistent with previous studies suggesting genetic diversity and abundance can vary amongst different B. pseudomallei STs and that there is an overrepresentation of a few STs within the Darwin region6,9. Five STs have been shown to comprise 90% of the overall clinical and environmental isolate abundance in Darwin and consistent with this, three of these were the most frequently identified molecular types in this study (ST-36, ST-109 and ST-553)9.
Results from this study also indicate that ST diversity can vary based on additional factors supplementary to geographic sampling area and size. We identified a significant association with increased ST diversity at park sites. Moreover, the odds of isolating more than one ST type were higher when these park sites were directly adjacent to or part of a sport’s field complex. While this finding contradicted our original hypothesis that drain catchment areas would be higher in ST diversity, previous environmental studies have shown that there is an increased occurrence of B. pseudomallei associated with sport’s fields34. This may be a consequence of optimal year-round growth conditions with higher rates of irrigation and maintained grass in the dry season, since irrigation and grass have been shown to increase the occurrence of B. pseudomallei in the environment35–37. This trend could also be associated with dissemination of the bacterium via contaminated shoes, since sport’s fields are visited by people from across the city. In line with above results, our data also showed a significant decrease in ST diversity in the outer region of the city (Region 4) compared to more urban central areas of Darwin.
Populations of B. pseudomallei are ecologically established and individual strain types are typically found less than 50 linear kilometres from one another in the environment6,7,17. Despite this, distinct genetic populations of the bacterium have only been documented across larger geographical boundaries, including the robust partitioning of Asian and Australian isolates at Wallace’s Line7,12,13,38. To-date, the smallest physical separation of B. pseudomallei populations identified has been amongst isolates from the Northern Territory, Australia and north Queensland, Australia9. Using high resolution WGS data and SNP-based Bayesian analysis, our results demonstrate that unique genetic populations of B. pseudomallei exist on an exceptionally small scale in the environment. RhierBAPS analysis of survey isolates identified five distinct populations of B. pseudomallei, implying there is a clear genetic distinction even within our narrow survey radius. While most clusters contained several different ST types, RhierBAPS partitioning identified two clusters each exclusively comprising only one unique ST, ST-109 or ST-553. This suggests that these two ST’s are less genetically diverse and more distantly related to the other strains we identified.
Contrary to this, Bayes Cluster 1 contained nearly 70% of the 35 STs identified, indicating some level of relatedness and shared ancestry amongst the majority of strains. Isolates that grouped within this cluster were shown to occur more frequently at drain sites. The high number of STs within this cluster could indicate an increased rate of genetic recombination at drainage catchment areas possibly related to the accumulation of B. pseudomallei isolates from different geographical areas, further suggesting that drains act as mechanisms of bacterial dissemination and dispersal in the environment. While these results were at odds to our previous findings indicating increased ST diversity at park sites, the latter association was observed at the site level while overall, there was a higher number of unique STs identified for drain soil isolates recovered. It is likely this trend would have been more significant had we cultured a higher percentage of qPCR-positive drain waters. However, direct PCR extraction post-enrichment has been demonstrated to be the most sensitive technique for the detection of B. pseudomallei in the environment compared to less-sensitive standard culture methods39,40. As a consequence of the high rate of qPCR-positive and low number of culture-positive drain water samples, the true ST diversity at drain sites was likely underestimated. Improved methods of culture detection and isolation of B. pseudomallei from water are crucial for future environmental surveys of the bacterium as determination of STs has not been possible from qPCR-positive culture-negative samples (data not shown).
Additionally, results from this study indicate that spatial clustering of B. pseudomallei populations can occur over a remarkably restricted geographical area, particularly for some B. pseudomallei genotypes. Three of the four common STs examined as part of the study were highly localised in the urban Darwin environment. While we identified no spatial clustering of ST-36, it is one of the Darwin region’s most frequently isolated molecular types and has been shown to be a significant source of melioidosis infection in the region since the Darwin Prospective Melioidosis Study (DPMS) began in 19895,6,9. We also found significantly more ST-36 in soil compared to water, collectively suggesting that the ST is well-established in the urban Darwin environment and this may be why we observed no spatial clustering for the strain. Interestingly, while ST-109 is also one of the region’s most frequently identified and widely dispersed molecular types, particularly in rural Darwin which was not included in this study, we observed a significant cluster within the city urban centre6,9,17. Since construction and earthwork projects have substantially increased in urban central Darwin in recent years, soil disturbance may have caused the strain to resurface and spread throughout the urban area. As we identified a significant association with ST-109 and water, it is possible the ST has been spreading throughout the urban centre via drainage areas and surface water runoff following periods of heavy rainfall.
Unlike ST-36 and ST-109, ST-327 and ST-553 have both become more common in the Darwin region recently in comparison to earlier years. For example, though first isolated from a Darwin melioidosis patient in 1990, ST-553 was observed rarely over the next 20 years. However, following the heavy 2009/2010 La Niña wet season, ST-553 has become one of the region’s most frequently isolated strains both in clinical patients and the environment (unpublished data)9, with more limited orthologous SNP diversity observed amongst isolates in comparison to more established, widely dispersed STs19,23. Moreover, results from this study also demonstrate that environmental ST-553 clustering around the north-eastern suburbs (Region 3) matched the residence of clinical cases with ST-553. While it is usually not known specifically where the clinical cases acquired B. pseudomallei, this result suggests infection may have taken place at or near their residential address. Moreover, the observation that ST “hot spot” areas and limited ST diversity in a region are reflected by a limited ST diversity of clinical isolates in that area supports the notion of there being no succinct subset of environmental B. pseudomallei strains capable of causing disease6,9. Despite this, while the presumptive location of B. pseudomallei infection for clinical cases of melioidosis cases can often be speculated utilising detailed patient epidemiological history and residential address, source attribution remains subjective. Additionally, significant correlation of clinical and environmental isolates did not occur for our other three common STs. Thus, further analysis examining the geographical associations and genomic similarities between clinical and environmental isolates belonging to the same STs are needed in the future. This knowledge will allow for better understanding of melioidosis source attribution in Darwin and may help to develop public health measures mitigating against the infection in other endemic regions.
Methods
Environmental sites and sample collection
Forty-two drains and 45 public park areas across urban Darwin (12.5°S) were selected for the study. Apart from stratifying sites across urban Darwin, selection of drains was also based upon site accessibility. The choice of park sites ensured that 3–5 sites were surveyed across all urban Darwin suburbs. All locations chosen for the survey were managed by Darwin City Council and site approval was granted prior to the survey commencing.
Darwin is a tropical savannah environment with distinctive wet and dry seasons. During the wet season the region receives an average annual rainfall of approximately 1,700 mm (http://www.bom.gov.au/climate/data/). For drain sites, samples were collected over two sampling rounds: one round was during the 2016 build-up (Oct-Nov) to capture storm-water runoff after the first major storms of the season (total rainfall accumulated since start of the wet season <200 mm) and the second round at the end of the 2017 monsoonal wet season (Feb-March, total accumulated rainfall since start of the wet season approximately 1,100 mm) (http://www.bom.gov.au/climate/data/). Three water samples were collected from each site using one litre collection bottles attached to bottle holders where water was present (dry season; samples n = 47 (19 sites), wet season; n = 108 (36 sites)). Five soil samples were also collected in the second sampling round from areas surrounding the drain (n = 210). Where possible, soils were spaced 10 m apart and taken from 30 cm depth41.
For park sites, ten soil samples each were collected from 45 sites (n = 450) at the end of the 2017 monsoonal wet season (Feb-March) using a fixed interval grid sampling approach with samples spaced 10 m apart and at 30 cm depth41. GPS coordinates were recorded at each site using a Garmin GPS device (Garmin eTrex30).
Environmental sample processing and confirmation
Culture of B. pseudomallei from soil and water was carried out using methods previously developed by Menzies School of Health Research41–43. Briefly, samples were enriched in Ashdown’s broth containing colistin (50 mg/L) and incubated at 37 °C aerobically for two and seven days. Enriched broth was plated onto Ashdown’s agar with gentamicin (8 mg/L) and incubated for 48 hours and colonies resembling B. pseudomallei were sub-cultured onto Ashdown’s agar. A maximum of ten B. pseudomallei-suspected colonies were sub-cultured per sample to account for genetic diversity within a sample. DNA from suspected colonies was extracted using 10% Chelex-100 resin44. Direct molecular detection of B. pseudomallei from soil and water samples was simultaneously preformed using the PowerSoil DNA isolation kit (MoBio Laboratories, USA) after an initial enrichment step of 24 h in Ashdown’s broth as previously described39. Confirmation of B. pseudomallei was done using a well-validated real-time PCR assay targeting a 115-bp segment within the type three secretion system 1 (TTS1) gene specific to the bacterium39,45,46.
B. pseudomallei isolates included for WGS analysis
A total of 135 environmental B. pseudomallei strains isolated during the investigation were used in this study for WGS population analysis and geographical mapping (Supplementary Table S2). One B. pseudomallei soil isolate and one water isolate from each wet and dry season culture-positive drain site were initially selected for WGS (n = 40). From the parks, one B. pseudomallei soil isolate was selected from each culture-positive site (n = 35). All isolates were chosen at random. To examine genetic variation between sites, additional isolates cultured from all drain and park samples were screened by BOX-PCR and visualised via gel electrophoresis using methods previously described by Menzies School of Health Research47. One isolate was selected from every culture-positive sample within a site (min number of isolates per site, n = 1; max number of isolates examined per site, n = 9) and screened against the single site isolate already selected for WGS. All sample isolates within a site that had a different banding patterns to the primary WGS isolate were also sent for sequencing (n = 60).
Whole-genome sequencing and sequence type (ST) assignment
Genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit (Qiagen, Chadstone, Victoria, Australia) as previously described47. Isolates were sequenced at Australian Genome Research Facility Ltd. (Melbourne, Australia) using the Illumina HiSeq. 2500 platform (Illumina, Inc., San Diego, CA). Multi-locus sequence typing (MLST) assignment of B. pseudomallei environmental soil and water isolates48 (n = 135) was assigned from WGS data in silico using the Bacterial Isolates Genome Sequence database (BIGSdb) tool accessible on the B. pseudomallei MLST website (http://pubmlst.org/bpseudomallei/)49.
Genome assembly and phylogenetic reconstruction
Orthologous core biallelic single-nucleotide polymorphisms (SNPs) and insertion and deletion events (InDels) were identified from WGS data using Genome Analysis Toolkit (GATK) in SPANDx v3.250. The closed Australian B. pseudomallei genome MSHR115351 was used as the reference for read mapping (N50:4,032,226 bp, contigs:2, total length:7,312,903 bp) and trees were rooted with MSHR0668, the most ancestral B. pseudomallei isolate identified to date based on a previous study52. A maximum-parsimony (MP) tree was constructed from 134,032 core orthologous SNPs and Indels using PAUP (v4.0a165)53 with 100 bootstrap replicates. A maximum-likelihood29 phylogenetic tree was inferred from WGS data using RAxML (v8.2.10)54 based on a normal model of sequence evolution using a gamma distribution. Trees were visualised in FigTree (v1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/) and manipulated using Interactive Tree of Life (iTOL v4) (https://itol.embl.de)55.
Hierarchical Bayesian clustering
A Bayesian approach was applied to spatially delineate and infer the genetic structure of Darwin B. pseudomallei isolates. Hierarchical clustering of the 135 genomes was done using RhierBAPS (version 1.1.2) implemented in R v.3.5.1 with a tree-independent approach using the core genome mapping alignment56–58. This method allows the population to be sub-divided into groups with closely related genetic backgrounds59. Clustering was performed until converging to a local optimum using two levels with k = 20 as the prior upper bound for the number of clusters. Higher levels of clustering were then performed based on the first result using k = 15 to k = 8. A value of k = 8 was chosen for both levels of clustering based on marginal likelihood values.
Statistical analysis
Statistical analyses were computed with Stata 14.0 (www.stata.com). Generalized estimating equation (GEE) and logistic regression models with robust standard errors clustered for site (67 sites comprising 135 WGS isolates) were used to analyse whether common STs and Bayesian Clusters were associated with sample (soil or water) or site type (drain or park) and region. The four urban Darwin regions were defined by location of water catchments and which catchment area survey sites drained into. An exchangeable intra-site correlation structures (ICC) was estimated for GEE models and odds ratios (ORs) for ST or Bayesian Cluster occurrence were calculated. A negative binomial model clustered for site was used to examine whether regions, sample or site types had a higher diversity of STs or Bayesian Clusters. Categorical variables were assessed using Fisher’s Exact tests and associations with significant environmental ST clusters and clinical isolates were examined using logistic regression analysis with data grouped by region. Clinical isolate location was assigned based on individual patient epidemiology and residential address. Statistical significance was determined using a p value less than 0.05.
Mapping of sites and spatial analysis of B. pseudomallei ST’s
Maps were created using ArcGIS 10.4.1 (ESRI) using GPS coordinates recorded at park and drain locations. Land unit shapefiles were obtained through the Northern Territory Government. The spatial structure of the four most frequently isolated urban Darwin STs was examined in ArcGIS implementing the Hot Spot Analysis (Getis-Ord Gi*) function with a fixed band distance. Results from the Gi* statistic were corrected for multiple testing and spatial dependence using the False Discovery Rate (FDR) correction method. Sites with GiZ scores >1.96 were considered significant at 95% confidence level (P < 0.05) and regarded as “hot” spots. GiZ scores < −2.58 indicated clustering of low values and were termed “cold” spots60.
Ethical approval
This study was approved by the Human Research Ethics Committee of the NT Department of Health and the Menzies School of Health Research (HREC 02/38). Sample site approval was obtained by the Darwin City Council before commencement of the study.
Supplementary information
Acknowledgements
We thank Darwin City Council for their technical advice and support in undertaking this investigation. Additionally, we thank Barbara Machunter for laboratory support, Erin Price and Derek Sarovich for genomics expertise and the microbiology staff at the Royal Darwin Hospital for their expertise in identifying B. pseudomallei and providing clinical isolates for this study. This study was supported by grants from the Australian National Health and Medical Research Council: grant numbers 1046812, 1098337 and 1131932 (The HOT NORTH initiative) and AR is supported by a Charles Darwin University International PhD scholarship.
Author contributions
The study was conceptualised by M.M., M.Ka., A.R., and B.J.C. Funding was acquired by B.J.C. Data collection was undertaken by A.R., M.M., G.H., and V.R. Formal analysis was done by A.R., M.K.l., M.Ka. and J.R.W. A.R. wrote the original draft, and M.Ka., B.J.C., M.M., J.R.W. revised and edited the manuscript.
Data availability
Raw sequence data from this study are available in the Short Read Archive in Bioproject PRJNA573745 (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA573745), biosample accessions SAMN12824487-SAMN12824621).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-62300-8.
References
- 1.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N. Engl. J. Med. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 3.Currie BJ, et al. Identification of melioidosis outbreak by multilocus variable number tandem repeat analysis. Emerg. Infect. Dis. 2009;15:169–174. doi: 10.3201/eid1502.081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarovich DS, et al. Whole-genome sequencing to investigate a non-clonal melioidosis cluster on a remote Australian island. Microb. Genom. 2017;3:e000117. doi: 10.1099/mgen.0.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS. Negl. Trop. Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapple SNJ, et al. Burkholderia pseudomallei genotype distribution in the Northern Territory, Australia. Am. J. Trop. Med. Hyg. 2016;94:68–72. doi: 10.4269/ajtmh.15-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz A, et al. Suspected cases of intracontinental Burkholderia pseudomallei sequence type homoplasy resolved using whole-genome sequencing. Microb. Genom. 2017;3:e000139. doi: 10.1099/mgen.0.000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Smet B, et al. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J. Clin. Microbiol. 2015;53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McRobb E, et al. Distribution of Burkholderia pseudomallei in northern Australia, a land of diversity. Appl. Environ. Microbiol. 2014;80:3463–3468. doi: 10.1128/aem.00128-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampton V, et al. Melioidosis in birds and Burkholderia pseudomallei dispersal, Australia. Emerg. Infect. Dis. 2011;17:1310–1311. doi: 10.3201/eid1707.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker A, et al. Groundwater seeps facilitate exposure to Burkholderia pseudomallei. Appl. Environ. Microbiol. 2011;77:7243–7246. doi: 10.1128/AEM.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson T, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC. Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chewapreecha C, et al. Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat. Microbiol. 2017;2:16263. doi: 10.1038/nmicrobiol.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price EP, et al. Whole-genome sequencing of Burkholderia pseudomallei isolates from an unusual melioidosis case identifies a polyclonal infection with the same multilocus sequence type. J. Clin. Microbiol. 2015;53:282–286. doi: 10.1128/JCM.02560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarovich, D. S. et al. Phylogenomic analysis reveals an Asian origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere1, 10.1128/mSphere.00089-15 (2016). [DOI] [PMC free article] [PubMed]
- 16.Dale J, et al. Epidemiological tracking and population assignment of the non-clonal bacterium, Burkholderia pseudomallei. PLoS. Negl. Trop. Dis. 2011;5:e1381. doi: 10.1371/journal.pntd.0001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rachlin, A. et al. A cluster of melioidosis infections in hatchling saltwater crocodiles (Crocodylus porosus) resolved using genome-wide comparison of a common north Australian strain of Burkholderia pseudomallei. Microb. Genom. 5, 10.1099/mgen.0.000288 (2019). [DOI] [PMC free article] [PubMed]
- 18.Smith S, Hanson J, Currie BJ. Melioidosis: An Australian Perspective. Trop. Med. Infect. Dis. 2018;3:27. doi: 10.3390/tropicalmed3010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parameswaran U, Baird RW, Ward LM, Currie BJ. Melioidosis at Royal Darwin Hospital in the big 2009–2010 wet season: comparison with the preceding 20 years. Med. J. Aust. 2012;196:345–348. doi: 10.5694/mja11.11170. [DOI] [PubMed] [Google Scholar]
- 20.Draper AD, et al. Association of the melioidosis agent Burkholderia pseudomallei with water parameters in rural water supplies in Northern Australia. Appl. Environ. Microbiol. 2010;76:5305–5307. doi: 10.1128/AEM.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McRobb E, et al. Melioidosis from contaminated bore water and successful UV sterilization. Am. J. Trop. Med. Hyg. 2013;89:367–368. doi: 10.4269/ajtmh.13-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McRobb E, et al. Tracing melioidosis back to the source: using whole-genome sequencing to investigate an outbreak originating from a contaminated domestic water supply. J. Clin. Microbiol. 2015;53:1144–1148. doi: 10.1128/JCM.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaestli M, et al. The association of melioidosis with climatic factors in Darwin, Australia: A 23-year time-series analysis. J. Infect. 2016;72:687–697. doi: 10.1016/j.jinf.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Baker AL, Warner JM. Burkholderia pseudomallei is frequently detected in groundwater that discharges to major watercourses in northern Australia. Folia. Microbiol. 2016;61:301–305. doi: 10.1007/s12223-015-0438-3. [DOI] [PubMed] [Google Scholar]
- 25.Rochelle-Newall E, Nguyen TM, Le TP, Sengtaheuanghoung O, Ribolzi O. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front. Microbiol. 2015;6:308. doi: 10.3389/fmicb.2015.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribolzi O, et al. Land use and soil type determine the presence of the pathogen Burkholderia pseudomallei in tropical rivers. Environ. Sci. Pollut. Res. 2016;23:7828–7839. doi: 10.1007/s11356-015-5943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann RE, et al. Rivers as carriers and potential sentinels for Burkholderia pseudomallei in Laos. Nat. Sci. Rep. 2018;8:8674. doi: 10.1038/s41598-018-26684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limmathurotsakul D, et al. Burkholderia pseudomallei Is spatially distributed in soil in Northeast Thailand. PLoS. Negl. Trop. Dis. 2010;4:e694. doi: 10.1371/journal.pntd.0000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corkeron M, Norton R, Nelson P. Spatial analysis of melioidosis distribution in a suburban area. Epidemiol. Infect. 2010;138:1346–1352. doi: 10.1017/S0950268809991634. [DOI] [PubMed] [Google Scholar]
- 30.Deiner, K., Fronhofer, E. A., Mächler, E., Walser, J.-C. & Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 7, 12544, 10.1038/ncomms12544https://www.nature.com/articles/ncomms12544#supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 31.Chuah CJ, Tan EKH, Sermswan RW, Ziegler AD. Hydrological connectivity and Burkholderia pseudomallei prevalence in wetland environments: investigating rice-farming community’s risk of exposure to melioidosis in North-East Thailand. Environ. Monit. Assess. 2017;189:287. doi: 10.1007/s10661-017-5988-1. [DOI] [PubMed] [Google Scholar]
- 32.Causse J, et al. Field and modelling studies of Escherichia coli loads in tropical streams of montane agro-ecosystems. J. Hydro-Env. rRes. 2015;9:496–507. doi: 10.1016/j.jher.2015.03.003. [DOI] [Google Scholar]
- 33.Currie BJ. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin. Respir. Crit. Care. Med. 2015;36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 34.Hill AA, et al. Melioidosis as a consequence of sporting activity. Am. J. Trop. Med. Hyg. 2013;89:365–366. doi: 10.4269/ajtmh.12-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaestli M, et al. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS. Negl. Trop. Dis. 2009;3:e364. doi: 10.1371/journal.pntd.0000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaestli M, et al. What drives the occurrence of the melioidosis bacterium Burkholderia pseudomallei in domestic gardens? PLoS. Negl. Trop. Dis. 2015;9:e0003635. doi: 10.1371/journal.pntd.0003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaestli M, et al. Out of the ground: aerial and exotic habitats of the melioidosis bacterium Burkholderia pseudomallei in grasses in Australia. Environ. Microbiol. 2012;14:2058–2070. doi: 10.1111/j.1462-2920.2011.02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace AR. On the zoological geography of the Malay Archipelago. Zool. J. Linn. Soc. 1860;4:172–184. doi: 10.1111/j.1096-3642.1860.tb00090.x. [DOI] [Google Scholar]
- 39.Kaestli M, et al. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl. Environ. Microbiol. 2007;73:6891–6897. doi: 10.1128/AEM.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knappik M, et al. Evaluation of molecular methods to improve the detection of Burkholderia pseudomallei in soil and water samples from Laos. Appl. Environ. Microbiol. 2015;81:3722–3727. doi: 10.1128/aem.04204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limmathurotsakul D, et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS. Negl. Trop. Dis. 2013;7:e2105–e2105. doi: 10.1371/journal.pntd.0002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayo M, et al. Burkholderia pseudomallei in unchlorinated domestic bore water, Tropical Northern Australia. Emerg. Infect. Dis. 2011;17:1283–1285. doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Currie BJ, et al. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg. Infect. Dis. 2015;21:2052–2054. doi: 10.3201/eid2111.141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res. Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-Y. [DOI] [PubMed] [Google Scholar]
- 45.Novak RT, et al. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 2006;44:85–90. doi: 10.1128/jcm.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaestli M, et al. Comparison of TaqMan PCR assays for detection of the melioidosis agent Burkholderia pseudomallei in clinical specimens. J. Clin. Microbiol. 2012;50:2059–2062. doi: 10.1128/JCM.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Currie BJ, et al. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC. Infect. Dis. 2007;7:68. doi: 10.1186/1471-2334-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godoy D, et al. Multilocus Sequence Typing and Evolutionary Relationships among the Causative Agents of Melioidosis and Glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003;41:2068–2079. doi: 10.1128/jcm.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC. Bionform. 2010;11:595–595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC. Res. Notes. 2014;7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson, S. L. et al. Whole-genome sequences of 80 environmental and clinical isolates of Burkholderia pseudomallei. Genome. Announc. 3, 10.1128/genomeA.01282-14 (2015). [DOI] [PMC free article] [PubMed]
- 52.Price EP, et al. Phylogeographic, genomic, and meropenem susceptibility analysis of Burkholderia ubonensis. PLoS. Negl. Trop. Dis. 2017;11:e0005928. doi: 10.1371/journal.pntd.0005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilgenbusch, J. C. & Swofford, D. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinform. Chapter 6, Unit 6 4, 10.1002/0471250953.bi0604s00 (2003). [DOI] [PubMed]
- 54.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 55.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic. Acids. Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corander J, Marttinen P, Siren J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC. Bioinform. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. RhierBAPS: An R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 2018;3:93–93. doi: 10.12688/wellcomeopenres.14694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Croucher NJ, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat. Genet. 2013;45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saxena R, et al. A spatial statistical approach to analyze malaria situation at micro level for priority control in Ranchi district, Jharkhand. Ind. J. Med. Res. 2012;136:776–782. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data from this study are available in the Short Read Archive in Bioproject PRJNA573745 (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA573745), biosample accessions SAMN12824487-SAMN12824621).