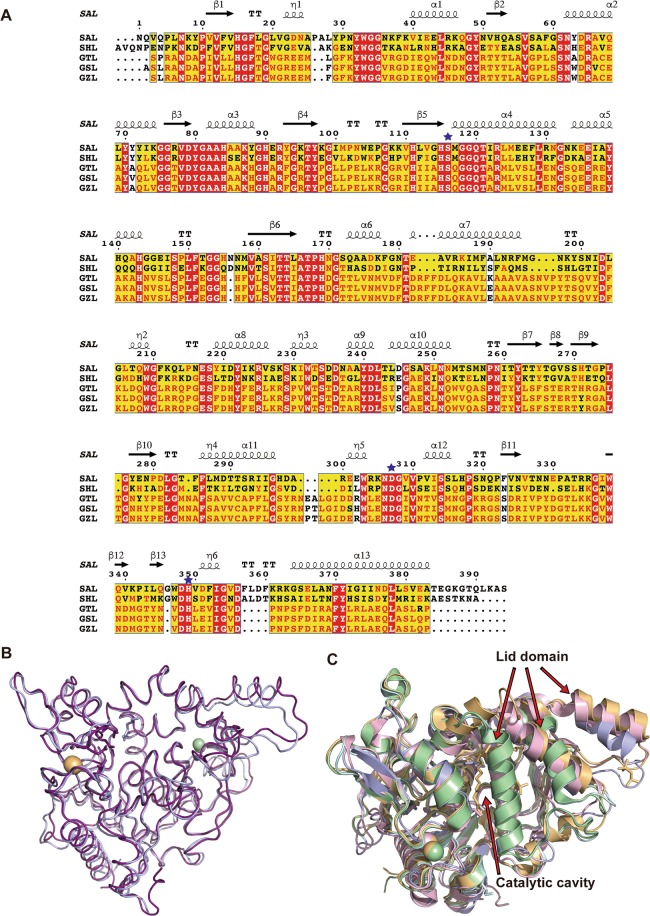
Figure 3.
(A) Sequence alignments of SAL, SHL, GTL, GSL, and GZL. Multiple sequence alignment of SAL and these structural homologous lipases was performed with ClustalW, followed by the ESPRIT program. The secondary structure elements of SAL are given above their corresponding sequences. Arrows and springs above the sequences indicate β-strand and helical conformations, respectively. The catalytic triad residues are marked with an asterisk. (B) Superimposition of SAL and SHL structures are shown in blue and pink, respectively. (C) Structural comparison of SAL with other homologous lipases. SAL, SHL, and GTL are shown in their open-lid form in blue, pink, and orange, respectively. GSL and GZL are shown in the closed form in green and cyan, respectively. Fatty acid fragment molecules are shown as sticks. The lids of GSL and GZL overlap with the position where fatty acids are localized in SAL.