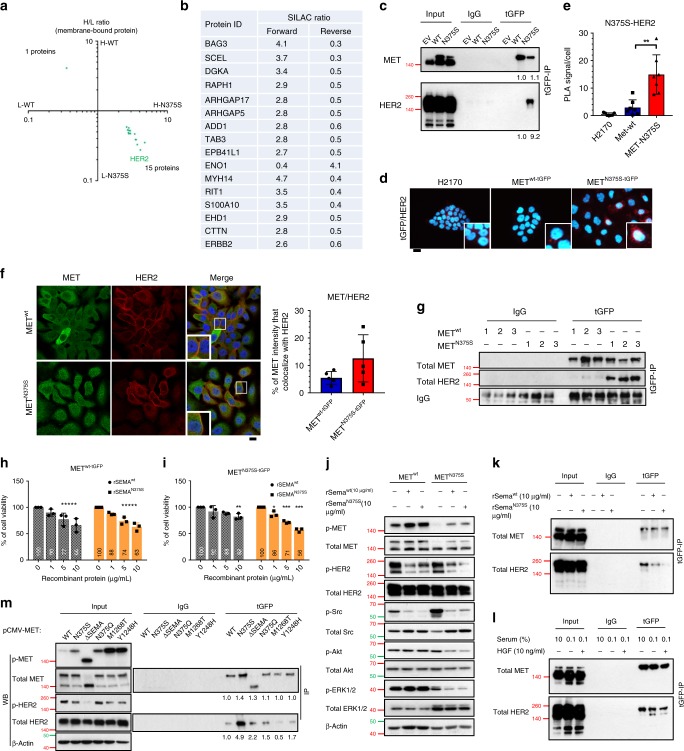
Fig. 4. The Sema domain of MET is critical for dimerization with HER2.
a, b Stable isotope labeling with amino acids in cell culture (SILAC) was performed to identify novel binding partners. METwt-tGFP and METN375S-tGFP cells were labeled with heavy (H) and light (L) amino acids. The cutoff values for SILAC ratios, after normalizing with MET expression, were set at (>2, <0.5) respectively. a Scatter plot of transformed METwt/METN375S ratios of membrane-bound proteins. Both axes represent METwt(H)/METN375S(L) and METwt(L)/METN375S(H) ratios, respectively. b List of various membranous proteins identified in SILAC analysis found to be associated with METN375S. c Interaction of ectopic MET and endogenous HER2 in H2170 METwt-tGFP and METN375S-tGFP cells was detected with immunoprecipitation and immunoblotting. Left, input controls. Total MET and HER2 band intensities, normalized to input controls and relative to METwt, are shown below (n = 5). d, e Detection of MET/HER2 co-localization (red) in EV, METwt-tGFP, and METN375S-tGFP cells with proximity ligation assay (PLA). Representative images are shown (d), with the PLA signals quantified (e) and expressed as the number of signals/cell ± SD (n > 3). Scale bar, 20 µm. f Representative confocal microscopy images of MET (Alexa-488; green) and HER2 (Alexa-594; red) in isogenic H2170 clones (n = 2). DAPI (blue) was used as nuclear counter stain. Co-localized proteins appeared in yellow. The smaller panels are detailed views of the outlined (white) squares in the respective images. Scale bar, 20 µm. The total MET fluorescence that co-localized with HER2 signal in each variant was tabulated on the right, data presented as mean ± SD (five fields in one representative experiment). g MET/HER2 interaction in H2170 METwt-tGFP and METN375S-tGFP tumors shown in Fig. 2g was detected with immunoprecipitation and immunoblotting. IgG was used as a loading control. h–k The role of Sema domain in METN375S-tGFP cells was examined with recombinant Sema proteins, wild-type, rSemawt; N375S mutant, rSemaN375S. Cell viability of METwt-tGFP (h) and METN375S-tGFP (i) cells after treatment with 1, 5, 10 µg/ml of rSemawt or rSemaN375S for 72 h, presented as mean ± SD (n = 3). Two-tailed Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001. j Immunoblots showing the total and phosphorylated expressions of MET, HER2, Src, Akt, and ERK1/2 in lysates of the indicated cell lines after treatment with 10 µg/ml rSema. β-Actin was used as a loading control. k MET/HER2 interaction in H2170 METN375S-tGFP cells was detected with immunoprecipitation and immunoblotting after treatment with 10 µg/ml rSema. Left, input controls. l MET/HER2 interaction in H2170 METN375S-tGFP cells was detected after serum starvation (0.1% FBS), or co-incubated with HGF (0.1% FBS + 10 ng/ml HGF). Left, input controls. m HEK293 cells were transfected with 1 µg of either pCMV6-EV-tGFP vector, MET-wt, N375S, ∆Sema, N375Q, M1268T, or Y1248H plasmid, together with pCMV6-ERBB2-DDK plasmid, for 24 h. MET/HER2 interaction in HEK293 cells was detected with immunoprecipitation and immunoblotting. Total MET and HER2 band intensities, relative to METwt, are shown below (n = 3). Left, input controls and phosphorylated proteins. β-Actin was used as a loading control.