Figure 1.
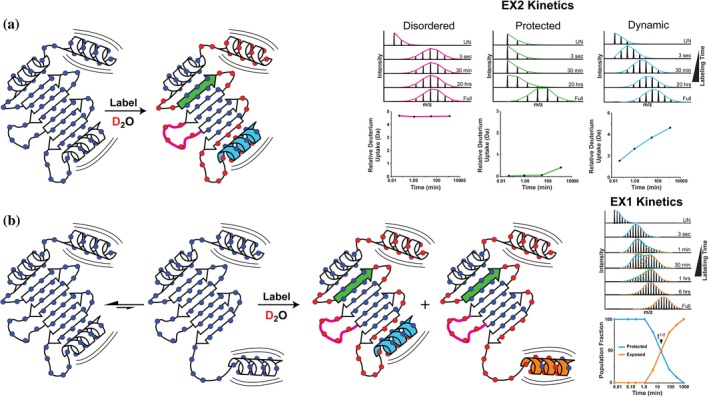
Protein structural dynamics and motion monitored by HDX‐MS. Continuous labeling HDX‐MS probes the accessibility of backbone amide hydrogens (blue circles) by their exchange with deuterium in solution. Under equilibrium conditions the majority of protein structural dynamics and motion manifests as “EX2 kinetics” (a) where deuterium is gradually incorporated across the protein backbone in a manner directly related to the local structural dynamics and changes in amide accessibility. Examples of peptide specific HDX‐MS data with representative mass spectra show the gradual incorporation of deuterium over time. Peptide segments in highly structured regions with strong hydrogen bonding networks (green beta sheet) take up deuterium much slower than regions with exposed and accessible amides (pink loop) and those undergoing dynamic motions (blue helix) that occur faster than the labeling rate. (b) When these dynamic structural changes are slower than the labeling rate (e.g., a reversible interconversion or conformational change) they produce unique and resolvable HDX states that manifest as “EX1 kinetics.” Here the protein reversibly interconverts between conformations with a protected (blue) or exposed (orange) helical domain. Analysis of the mass spectra reveals the equilibrium distribution of these two states and their respective half‐lives
