Figure 3.
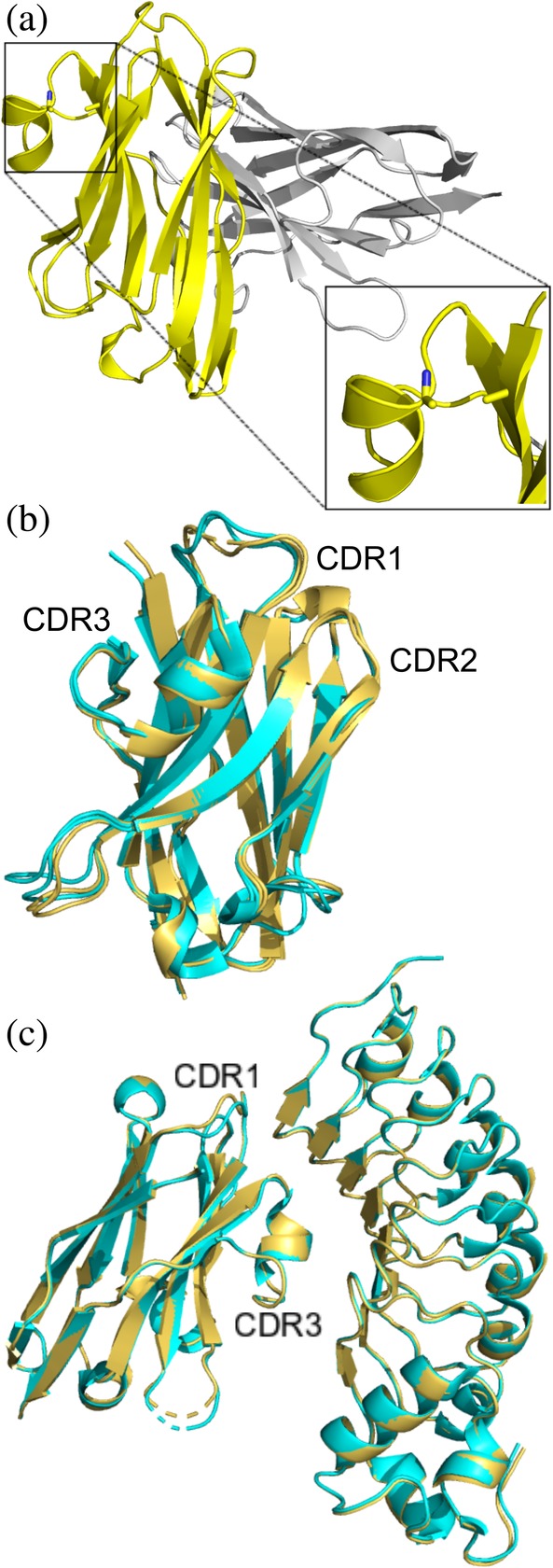
X‐ray structure of unliganded R303C33A/C102A and in complex with InlB249 (a) Structure of unliganded R303C33A/C102A (PDB code: http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6U14) contains a dimer in the asymmetric unit. The C33A/C102A double mutation is clearly visible (highlighted in the zoomed in box). (b) Overlay of R303C33A/C102A (yellow ribbon structure) with wild type R303 (cyan, PDB code: http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6DBA) showing only subtle changes in the structure of CDR1. (c) Structure of R303C33A/C102A in complex with InlB249 (yellow, PDB code: http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6U12) aligned with the structure of R303 in complex with InlB249 (cyan, PDB code: http://bioinformatics.org/firstglance/fgij//fg.htm?mol=6DBF). The CDR loops and VHH‐antigen interactions are nearly identical in the two structures
