Fig. 7.
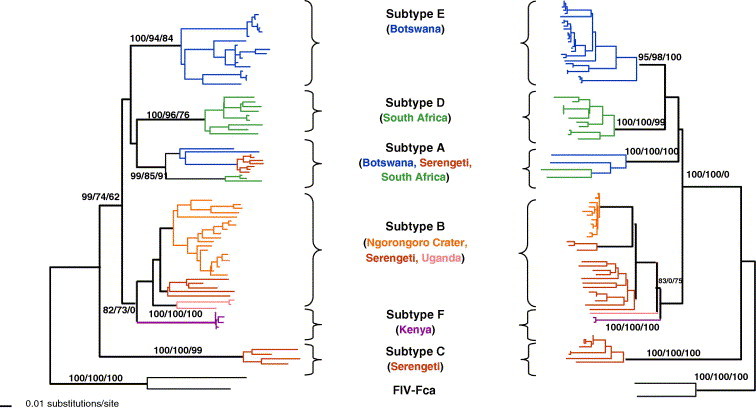
Phylogenetic tree for FIV pol-RT (520 bp included in analysis) and gag (400 bp) nucleotide sequences isolated from lion populations sampled in Fig. 6. Shown here is the single maximum likelihood (ML) tree. Maximum parsimony (MP), and minimum evolution (ME; Neighbor joining based on the Tajima Nei algorithm for distance measure) trees gave similar topologies. Bootstrap values are included at all nodes with bootstrap support (ML/ME/MP). Taxa are colored by the country of origin of the lion sampled. FIV–FCA is the outgroup strain of domestic cat FIV.
