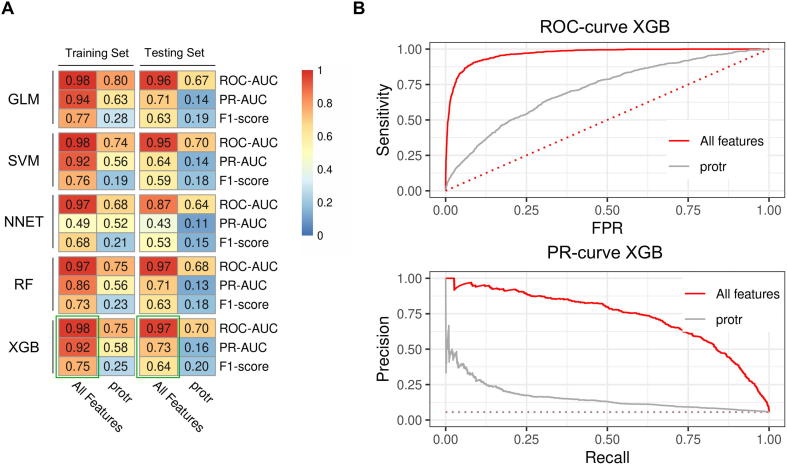
Fig. 6.
Machine learning using a broad range of features leads to high classification performance in human. (A) Heatmaps showing accuracy metrics for the performance evaluation of essential gene classification in human. Five ML approaches were used (Generalised linear model [GLM], Support-Vector Machines [SVM)], Neural Network [NNET], Random Forests [RF], and Extreme Gradient Boosting [XGB]). The performance was measured for the training and testing sets. Features were generated following our new approach including a broad range of features (All Features) and the performance was benchmarked to protein sequence features only (protr). The algorithm with the best performance (highest harmonic mean) was XGB, indicated by a green frame. (B) Receiver operating characteristic and Precision-Recall curves from XGB classifier measured on the testing sets. Using all features (red) yielded distinctively better results compared to the results from the protein sequence features only (grey). Results from a random classification is indicated by dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)