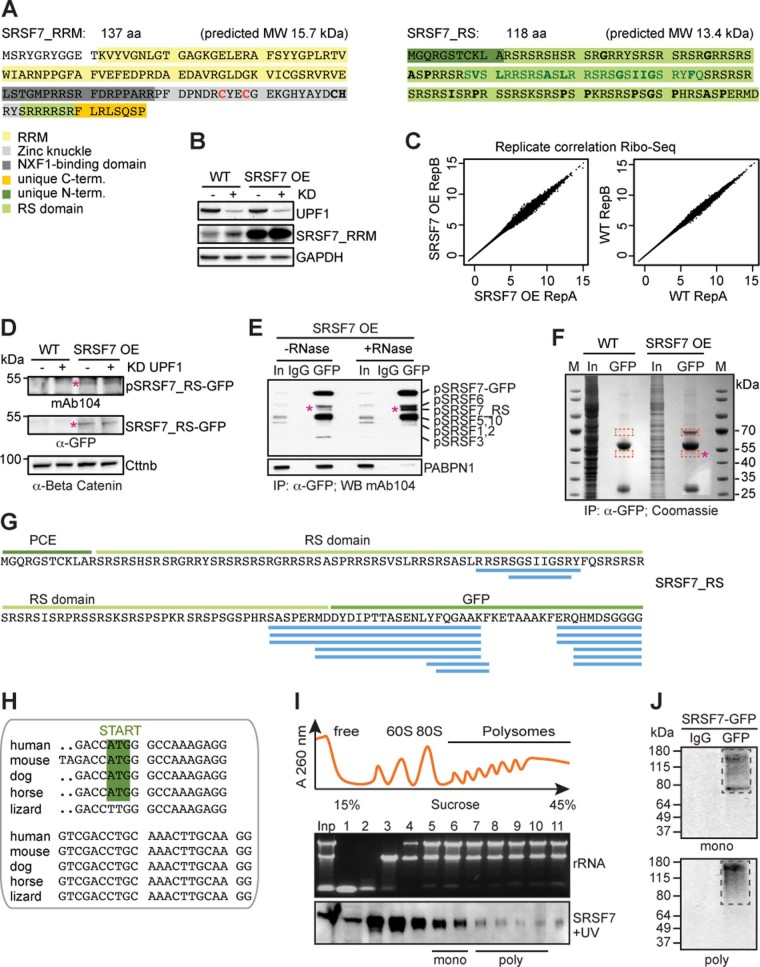
Extended Data Fig. 2. The SRSF7-PCE isoform is translated into two distinct truncated SRSF7 proteins.
a, Sequence and predicted molecular weight (MW) of the SRSF7_RRM and SRSF7_RS isoforms. SRSF7 protein domains are highlighted with the indicated colors. Hydrophobic amino acids within the RS domain are indicated in bold. The deleted 27aa stretch (see Fig. 6) is indicated in dark green. The mutated cysteine residues are indicated in red. b, SRSF7_RRM level in WT and SRSF7 OE cells upon knockdown (KD) of UPF1. The WB membrane was probed with α-SRSF7 and α-UPF1 antibodies. GAPDH was used as loading control. c, Ribo-Seq was performed from WT (Ctrl) and SRSF7 OE cells in two replicates. Scatter plot of Rlog-transformed raw Ribo-Seq reads from both replicates. d, WB analysis of WT and SRSF7 OE cells upon knockdown of UPF1. The blot was probed with mAb104 (anti-phospho-RS) and α-GFP. α-Beta-Catenin (Cttnb) was used as loading control. e, Co-IP of purified SRSF7-GFP probed with mAb104 to verify the presence of phosphorylated SRSF7_RS. PABPN1 was used to control for RNase treatment. IgG – unspecific antibody control, In – Input. f, Coomassie-stained SDS-PAGE of a stringent IP to purify the SRSF7_RS-GFP isoform for MS analysis. Cut bands are indicated in orange squares. M – marker g, Identified high-confidence peptides (FDR<0.01) mapping to the PCE, the RS domain and GFP. h, Alignment of PCE 3’ends indicate conservation of the in-frame START codon in mammals. i, iCLIP from polysomal fractions (piCLIP). Polysome profile after UV crosslinking of SRSF7-GFP to RNA in P19 cells. Indicated fractions were pooled to obtain monosomal (5+6) and polysomal (7–10) fractions. A representative rRNA gel and α-GFP WB are shown below (n=3). j, Protein-RNA complexes were immunopurified under stringent conditions from pooled fractions using α-GFP antibodies. RNA-protein complexes were undetectable when unspecific antibodies (IgG) were used.