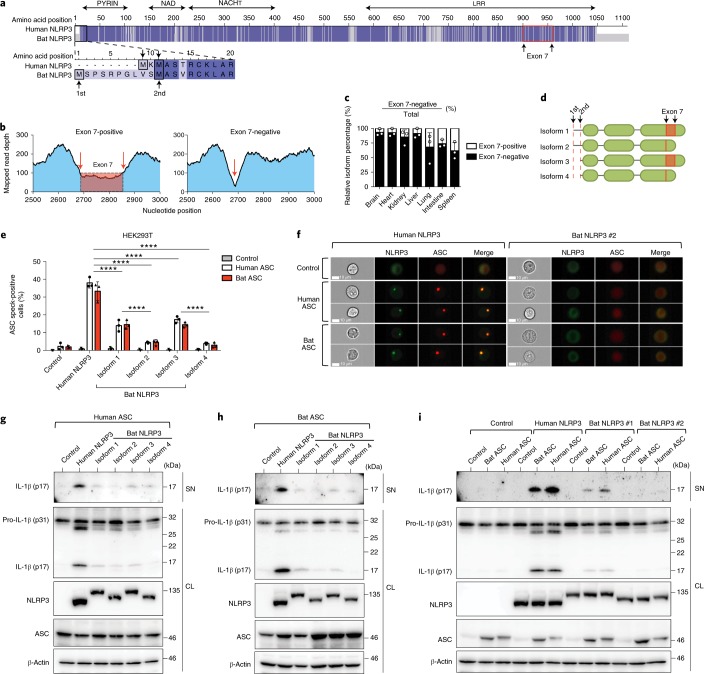
Fig. 3. The function of all four bat NLRP3 isoforms, but not ASC, is reduced.
a, Amino acid sequence alignment of human and bat (P. alecto) NLRP3, with identical residues highlighted in dark blue and mismatches in light blue. Black boxes, alternative translation start sites. Red box, boundaries of exon 7. b, Read-mapping analysis of two splice variants (exon 7-positive/negative) by RNA–seq read counts of the P. alecto splenocytes subsets. Arrows, exon–exon junctions. c, Percentage of exon 7-positive or exon 7-negative isoform in P. alecto tissues measured by qPCR. d, Schematic representation of four bat NRLP3 isoforms. e, Quantification of ASC specks by ImageStream in HEK293T cells stably expressing mPlum (control), human or bat ASC–mPlum, transfected with mCitrine (control), human or bat NLRP3–mCitrine. f, Representative images from e. NLRP3–mCitrine (green); ASC–mPlum (red). Scale bars, 10 μm. g–i, Immunoblot analysis of IL-1β cleavage in cell lysates and supernatant in HEK293T cells stably expressing mPlum (control), human or bat ASC–mPlum, transfected with mCitrine (control), human or bat NLRP3–mCitrine, plus human pro-caspase-1 and mouse pro-IL-1β. NLRP3, NLRP3–mCitrine stained with anti-GFP; ASC, ASC–mPlum stained with anti-mPlum; SN, supernatant; CL, cell lysate. ****P < 0.0001 by two-way ANOVA with Bonferroni’s multiple comparisons test (e). Exact P values are provided in Supplementary Table 5. Data are presented as mean + s.d. of three biological replicates (c) or three independent experiments (e), or representative of three independent experiments (f–i).