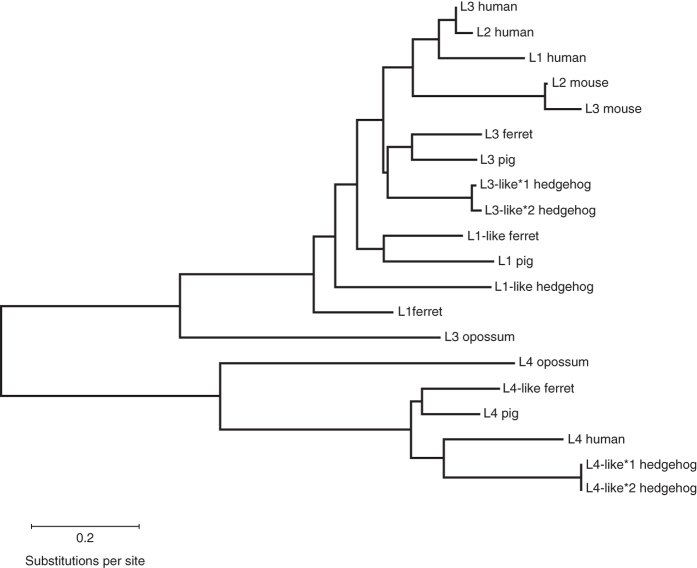
Figure 4. Phylogenetic tree of IFN-λ based on actual or predicted protein sequences.
We selected species so that they represented different branches of the mammalian clade. We selected one species of marsupials (gray short-tailed opossum; representing early mammalian evolution) and three representative species from the superorder Laurasiatheria (ferret, pig, hedgehog) as well as mouse and human, which we deal with in this review and represent the two major branches of the superorder Euarchontoglires. For the analysis, we kept the protein names as found in the NCBI database and only added numbers to proteins that had redundant names. The sequences were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/), and the tree was generated by the Maximum Likelihood method based on the JTT matrix-based model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.