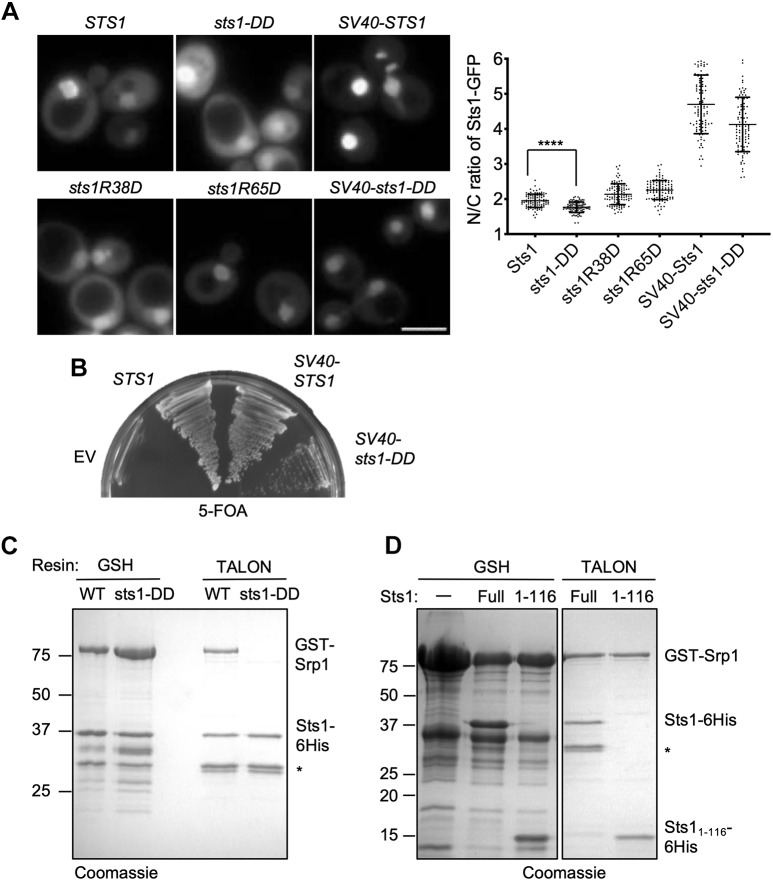
Fig. 2.
Nuclear localization is essential for Sts1 function. (A) Wild-type MHY500 yeast was transformed with p415GPD-based plasmids expressing the indicated sts1-GFP fusion alleles. ‘SV40’ denotes the cNLS from SV40T antigen fused to the N-terminus of the test protein. The transformants were grown at room temperature prior to imaging by fluorescence microscopy in mid-log phase. Three replicates of at least 100 cells were counted (right panel). A t-test was used to determine statistical significance of differences in localization (****P<0.0001). Scale bar: 5 μm. (B) Viability assay. MHY9580 cells (sts1Δ, pRS316-STS1) were transformed with the p415GPD-based alleles noted and grown at room temperature on 5-FOA to select against the original cover plasmid. (C) Sts1–Srp1 complex formation depends on the Sts1 bipartite NLS. The indicated recombinant proteins were co-expressed in E. coli, and binding was determined based on co-purification on a GST-binding glutathione resin or polyHis-binding TALON resin followed by SDS-PAGE. (D) The indicated recombinant proteins were co-expressed in E. coli, and binding was determined based on co-purification on either GSH beads or TALON beads. *Proteolytic fragments derived from Sts1–6His.