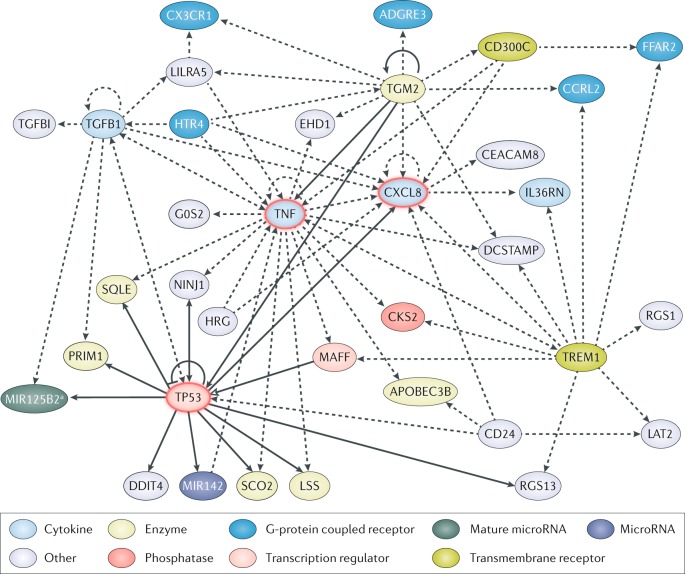
Fig. 2. Network of candidate genes for predicting patient mortality identified through prognostic enrichment strategies.
The community approach to mortality risk prediction in a sepsis30 study identified 65 risk-predicting genes based on publicly available gene expression data from adults, children and neonate with sepsis. Similarly, during the initial discovery phase of the PERSEVERE biomarkers32 used for prognostic enrichment, 80 genes associated with poor outcome were selected based on transcriptomic data from a cohort of paediatric patients with septic shock. An analysis of the genes identified in these two studies revealed 11 genes in common: adhesion G protein-coupled receptor E3 (ADGRE3), CD24 molecule (CD24), carcinoembryonic antigen-related cell adhesion molecule 8 (CEACAM8), CDC28 protein kinase regulatory subunit 2 (CKS2), C-X3-C motif chemokine receptor 1 (CX3CR1), DNA damage inducible transcript 4 (DDIT4), G0/G1 switch 2 (G0S2), C-X-C motif chemokine ligand 8 (CXCL8, also known as IL-8), MAF bZIP transcription factor F (MAFF), regulator of G protein signalling 1 (RGS1) and transforming growth factor beta induced (TGFBI). The network was generated by uploading these 11 genes to the Ingenuity Pathway Analysis platform. Solid lines represent direct interactions and dashed lines represent indirect interactions; solid arrowheads can represent activation, modification and phosphorylation among others (for more details see Ingenuity Pathway Analysis legend), open arrowhead represents translocation and bar-headed arrow indicates that the gene inhibits and acts on its target. The gene network contains three highly connected central nodes relevant to the pathobiology of sepsis and inflammation: tumour necrosis factor (TNF), CXCL8 and tumour protein p53 (TP53). APOBEC3B, apolipoprotein B mRNA editing enzyme catalytic subunit 3B; CCRL2, C-C motif chemokine receptor like 2; CD300C, CMRF35-like molecule 6; DCSTAMP, dendrocyte expressed seven transmembrane protein; EHD1, EH domain containing 1; FFAR2, free fatty acid receptor 2; HRG, histidine-rich glycoprotein; HTR4, 5-hydroxytryptamine receptor 4; IL36RN, interleukin 36 receptor antagonist; LAT2, linker for activation of T cells family member 2; LILRA5, leukocyte immunoglobulin-like receptor A5; LSS, lanosterol synthase; MIR142, microRNA 142; NINJ1, ninjurin 1; PRIM1, DNA primase subunit 1; RGS13, regulator of G protein signalling 13; SCO2, SCO cytochrome c oxidase assembly protein 2; SQLE, squalene epoxidase; TGFB1, transforming growth factor beta 1; TGM2, transglutaminase 2; TREM1, triggering receptor expressed on myeloid cells 1. aRefers to the mature sequence miR-125b-2–3p, encoded in the micro RNA-125b-2 (MIR125B2) stem loop sequence, and includes other microRNAs with the seed sequence CAAGUCA.