Figure 4. Viral genome replication directed by long-distance interactions.
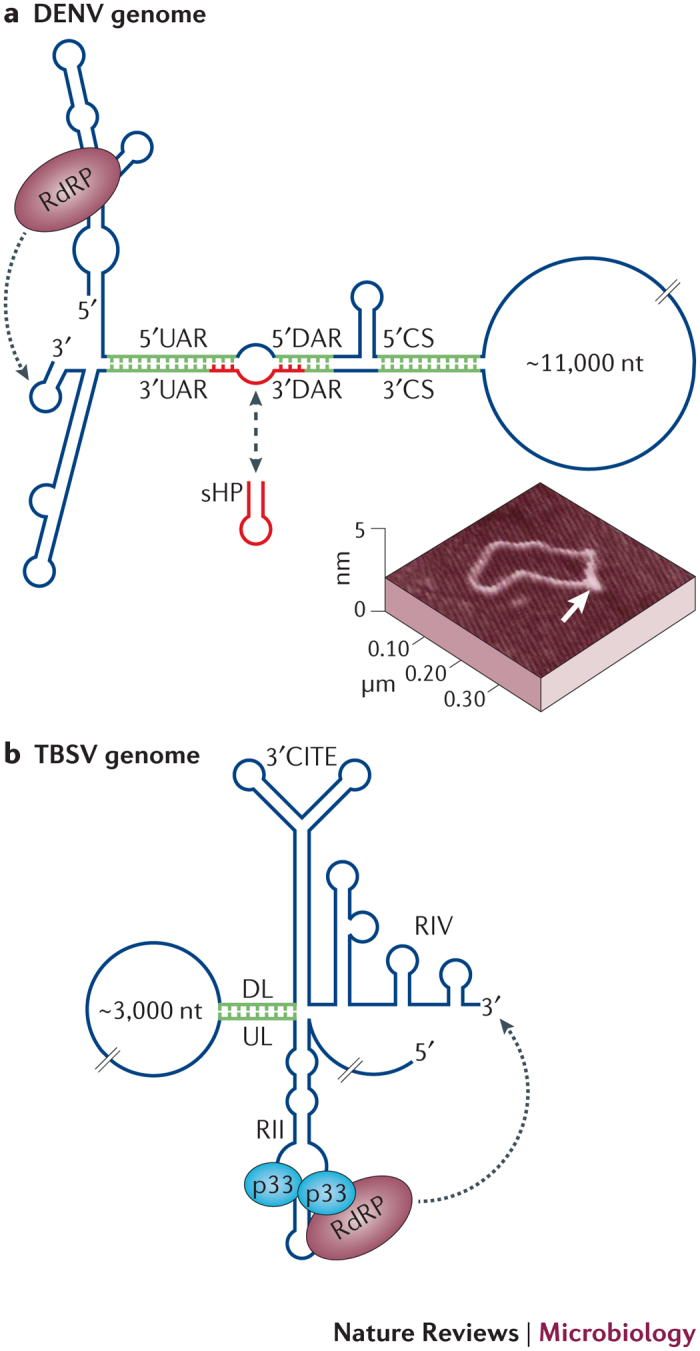
a | A simplified representation of relevant RNA secondary structures in the dengue virus (DENV) RNA genome. Genome replication requires three sets of long-distance interactions: 5′−3′ upstream of AUG region (UAR), 5′−3′ downstream of AUG region (DAR) and 5′ cyclization sequence−3′ cyclization sequence (CS). Formation of these interactions results in genome circularization, which repositions the 5′-bound RNA-dependent RNA polymerase (RdRp) to the 3′ end of the genome, where it initiates negative-strand synthesis. Interacting sequences are shown in green. The formation of the alternate regulatory structure small hairpin (sHP; shown in red) inhibits circularization. A circularized version of the genome, as observed by atomic force microscopy (AFM), is shown, with the 5′−3′ base-paired region denoted by a white arrow65. b | A simplified representation of the RNA secondary structures in the tomato bushy stunt virus (TBSV) RNA genome that are involved in genome replication. The 3′ cap-independent translational enhancer (3′ CITE) region is also included, as the downstream linker is located immediately upstream of it. The RdRp interacts with its auxiliary replication protein p33, which binds as a dimer to an internal RNA element, RII. A long-distance interaction that involves the upstream linker and downstream linker (UL–DL) sequences brings RdRp, which is associated with RII close to the 3′ end of the genome, where it can initiate negative-strand synthesis. The UL–DL interaction also mediates the formation of a RII–RIV RNA platform that facilitates the assembly of the viral replicase complex. The AFM image in part a is reproduced, with permission, from Filomatori, C. V. et al. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20, 2238–2249 (2006) © Cold Spring Harbor Laboratory Press.
