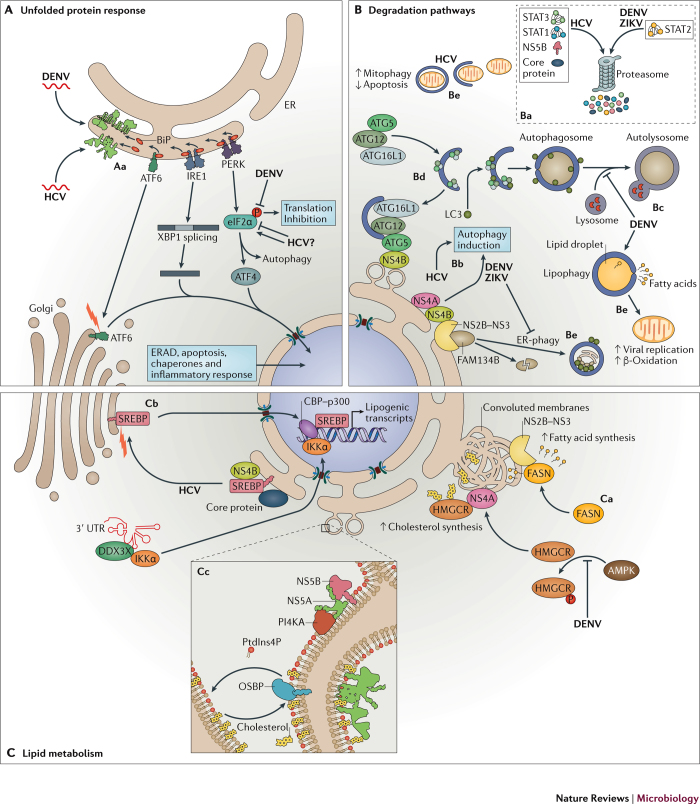
Figure 3. Cellular pathways co-opted by Flavivirus and Hepacivirus.
A | Expression of the viral proteins during hepatitis C virus (HCV) and dengue virus (DENV) infection recruits immunoglobulin heavy chain-binding protein (BiP), resulting in depletion of BiP from the endoplasmic reticulum (ER)-stress sensors cyclic AMP-dependent transcription factor ATF6-α (ATF6), serine/threonine-protein kinase/endoribonuclease IRE1 and eukaryotic translation initiation factor 2-α kinase 3 (PERK). Aa | These BiP-less sensors become activated to induce the unfolded protein response (UPR) pathway. HCV and DENV have developed strategies to manipulate UPR pathways to promote virus replication (see main text for details). B | Manipulation of the cellular degradation pathways. Ba | HCV, DENV and Zika virus (ZIKV) infection induce proteasome-mediated degradation of innate signalling proteins to dampen the antiviral immune response. Bb | Viral infection triggers autophagy and alters the autophagy flux. Bc | For instance, established DENV infection suppresses the autophagic flux, blocking the fusion between autophagosomes and lysosomes. Bd | HCV non-structural proten 4B (NS4B) interacts with autophagy protein 5 (ATG5) and recruits the autophagosome membrane elongation complex (ATG5–ATG12–ATG16L1) to the membranous web, contributing to its formation. Be | In addition to general autophagy, selective autophagic pathways are hijacked during Flaviviridae infection (see main text for details). C | Subversion of lipid homeostasis. Ca | DENV serine protease NS3 recruits fatty acid synthase (FASN) to replication compartments to stimulate lipid biosynthesis. De novo synthesized fatty acids are incorporated into replication organelles (ROs). 3-Hydroxy-3-methylglutaryl-CoA reductase (HMGCR) is recruited to DENV ROs, where it colocalizes with NS4A. Inactivation of the protein kinase 5′-AMP-activated protein kinase (AMPK) during DENV infection enhances HMGCR enzymatic activity, which results in higher cholesterol levels within ROs. Cb | During HCV infection, both viral proteins and regulatory elements within the 3′ UTR of the viral genome activate the transcription of lipogenic genes through sterol regulatory element-binding protein (SREBP)-mediated pathways (see main text for details). Cc | HCV NS5A and NS5B recruit and stimulate phosphatidylinositol 4-kinase-α (PI4KA) activity, thus increasing the local concentration of phosphatidylinositol-4- phosphate (PtdIns4P). Lipid transfer proteins such as oxysterol-binding protein 1 (OSBP) bind to PtdIns4P-rich membranes and release cholesterol in exchange for PtdIns4P. ATF4, cyclic AMP-dependent transcription factor ATF-4; ATG12, ubiquitin-like protein ATG12; ATG16L1, autophagy-related protein 16-like 1; CBP, CREB-binding protein; DDX3X, ATP-dependent RNA helicase DDX3X; eIF2α, eukaryotic translation initiation factor 2 subunit 1; ERAD, ER-associated degradation; FAM134B, reticulophagy regulator 1; IKKα, inhibitor of nuclear factor-κB kinase subunit-α; LC3, microtubule-associated proteins 1A/1B light chain 3B (also known as MAP1LC3B); p300, histone acetyltransferase p300; STAT1, signal transducer and activator of transcription 1; STAT2, signal transducer and activator of transcription 2; STAT3, signal transducer and activator of transcription 3; XBP1, X-box-binding protein 1.