Figure 1. The miRNA and siRNA pathways of RNAi in mammals.
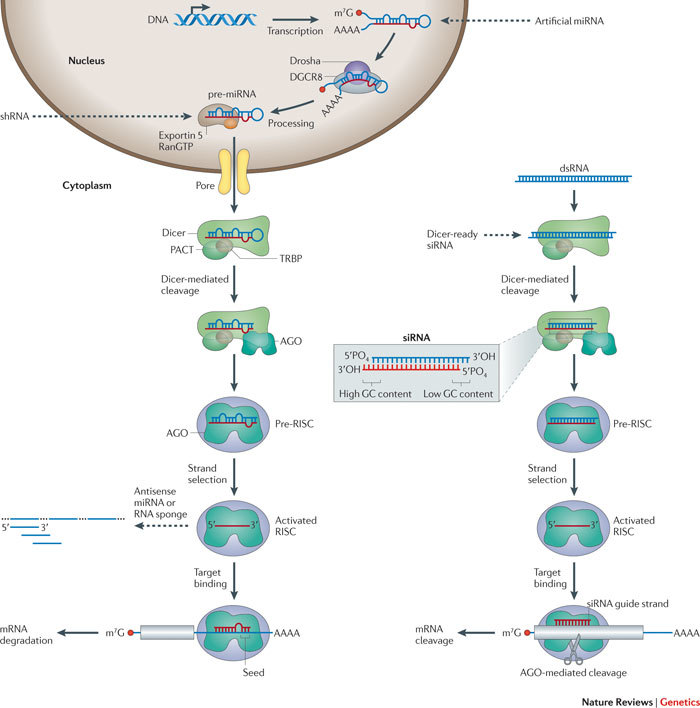
Primary microRNAs (pri-miRNAs) are transcribed by RNA polymerases156,157,158 and are trimmed by the microprocessor complex (comprising Drosha and microprocessor complex subunit DCGR8) into ~70 nucleotide precursors, called pre-miRNAs67,159,160 (left side of the figure). miRNAs can also be processed from spliced short introns (known as mirtrons)161. pre-miRNAs contain a loop and usually have interspersed mismatches along the duplex. pre-miRNAs associate with exportin 5 and are exported to the cytoplasm162,163, where a complex that contains Dicer, TAR RNA-binding protein (TRBP; also known as TARBP2) and PACT (also known as PRKRA) processes the pre-miRNAs into miRNA–miRNA* duplexes116,164,165. The duplex associates with an Argonaute (AGO) protein within the precursor RNAi-induced silencing complex (pre-RISC). One strand of the duplex (the passenger strand) is removed. The mature RISC contains the guide strand, which directs the complex to the target mRNA for post-transcriptional gene silencing. The 'seed' region of an miRNA is indicated; in RNAi trigger design, the off-target potential of this sequence needs to be considered. Long dsRNAs (right side of the figure) are processed by Dicer, TRBP and PACT into small interfering RNAs (siRNAs). siRNAs are 20–24-mer RNAs and harbour 3′OH and 5′ phosphate (PO4) groups, with 3′ dinucleotide overhangs3,166,167. Within the pre-RISC complex, an AGO protein cleaves the passenger siRNA strand. Then, the mature RISC, containing an AGO protein and the guide strand, associates with the target mRNA for cleavage. The inset shows the properties of siRNAs. The thermodynamic stability of the terminal sequences will direct strand loading. Like naturally occurring or artificially engineered miRNAs, the potential 'seed' region can be a source for miRNA-like off-target silencing. shRNA, short hairpin RNA.
