Figure 2. Architecture of eukaryotic membrane and viral fusion proteins compared with bacterial invasins.
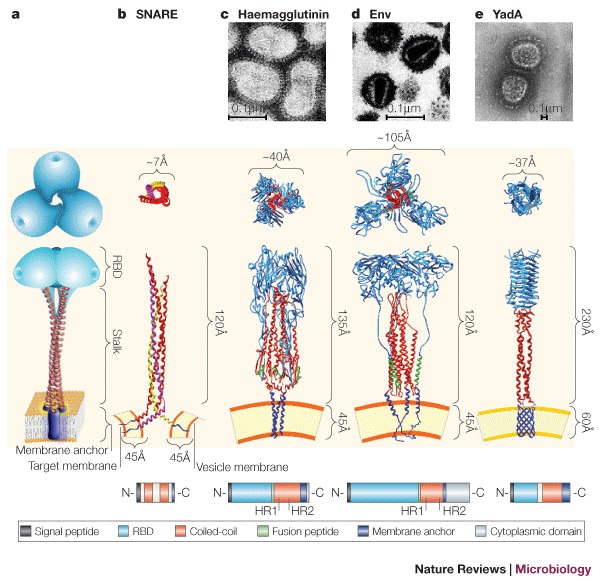
a | Schematic representation of the common trimeric structure consisting of a membrane anchor (purple), a central coiled-coil motif (stalk, red) and a globular receptor-binding domain (RBD, cyan). b | Partial three-dimensional structure of the coiled-coil domains of the synaptic SNARE complex. Synaptobrevin 2 (also known as VAMP2) (yellow) is anchored to the vesicle membrane. Syntaxin (pink) and SNAP25 (red) are anchored to the target membrane. These proteins are anchored to the cytoplasmic membrane by a hydrophobic α-helical C- terminal domain, which allows the protein to extend 120Å from the cell membrane. c | Structural representation of the influenza haemagglutinin (HA) trimer with the fusion peptide (green). The stalk is in its metastable pre-fusogenic state. The globular receptor-binding subunit HA1 is shown in blue, and the triple-stranded, coiled-coil fibrous region, known as HA2, is shown in red. d | The HIV envelope protein (Env) is composed of two non-covalently associated trimeric gp120 (red) and gp41 (blue) subunits. Here, gp120 is artificially fused to gp41. The fusion peptide, which is located in gp41, is depicted in green. In their monomeric form, both fusion proteins — haemagglutinin and Env — span the viral envelope with a single transmembrane α-helix near the C-terminus. e | Structural model of the trimeric Yersinia spp. YadA molecule anchored to the bacterial outer membrane through a β-barrel (blue). The anchor domain is represented by a porin-like outer membrane protein. Top–down views of the heterotrimeric SNARE complex, homotrimeric viral and bacterial fusion proteins (invasins), are shown. Structures are not to scale. The structures used to build the models have been obtained from the PDB protein databank (see the Online links box). Short domains for which coordinates were not available were built manually on the basis of secondary structure predictions. The anchor domain was modelled on the integral outer membrane protein (OmpA) from Escherichia coli (strain 1BXW), whereas the coiled-coil stalk was modelled on the structure of chicken cartilage trix protein (1AQ5). Transmission electron microscopy (TEM) of negatively stained influenza virus (c), HIV (d) and Yersinia spp. (e) are shown. The bottom panel is a schematic representation of the protein sequence architecture of the corresponding proteins. HR1 and HR2 are experimentally determined heptad repeats for haemagglutinin and Env. White boxes represent sequence portions with an undefined role. SNARE, soluble NSF (N-ethylmaleimide-sensitive fusion protein) accessory protein (SNAP) receptor. Panel c TEM courtesy of Linda M. Stannard, University of Cape Town, South Africa. Panel c (structural image) reproduced, with permission, from Ref. 98 © (2002) Elsevier Science. Panel d TEM courtesy of H. Gelderblom, Koch Institute, Berlin. Panel e TEM reproduced, with permission, from Ref. 60 © (2000) European Molecular Biology Organization.
