Abstract
Severe acute respiratory syndrome (SARS) took everybody by surprise. Its emergence was one of the most significant microbiological events of 2003. It challenged microbiologists to identify the aetiological agent and satisfy Koch's postulates — in so far as they ever can be met for a virus — in real time. Not only were the patients' respiratory secretions and the agents grown in cultured cells put under the microscope, but so were the actions of politicians. What lessons can we learn from SARS?
Main
The common cry uttered after crises is that 'lessons must be learned'. What lessons does SARS teach? The crisis is over, at least for the time being, but how certain can we be that the German philosopher Hegel was wrong when he said “what experience and history teach is this — that people and governments have never learned anything from history, or acted upon principles deduced from it”?
Before attempting to answer these questions using SARS as an appropriate example, it is necessary to consider the SARS virus itself and ask another question. How severe a test did SARS pose? Were we lucky? The answer is 'yes, we were'. This is qualified by the number of deaths that it caused; however, it is due, at least in part, to the properties of the virus. Most important is the low R0 value — the basic reproductive number — for the SARS virus, which represents the number of secondary infectious cases that are generated by an average infectious case in a susceptible population. The R0 value predicts the likelihood that an infectious agent will start an outbreak, the amount of transmission that can be expected in the absence of control measures and the ability of these control measures to reduce spread. Studies on the SARS outbreak in Hong Kong1 — after the exclusion of two 'super-spread' events where special circumstances allowed index cases to infect many individuals (at the Prince of Wales Hospital and at the Amoy Gardens estate) — gave an estimated R0 value of 2.7. This is much lower than for any other virus that is spread by the respiratory route; R0 values for such infections in England and Wales have been estimated to be 16–18 for measles, 10–12 for chickenpox, 11–14 for measles and 6–7 for rubella2.
Controlling virus spread
The low R0 value for SARS indicates that controlling the spread of infection should be easier to achieve than for many other respiratory viruses. The only other virus spread by the respiratory route with an R0 value approaching that of SARS is smallpox. For isolated pre-twentieth century populations with negligible immunity, Gani and Leach3 estimated the R0 value for smallpox to be 3.5–6.0 and, for 30 importations into Europe between 1958 and 1973, to be about 5.5 for community-acquired disease. Although an R0 value of this magnitude means that without control measures an outbreak would grow exponentially, in the case of smallpox outbreaks it was always possible to rapidly reduce the value of R0 during the progress of an outbreak. Maintaining the R0 value below 1 prevented the development of secondary cases; it was achieved by locating and isolating cases, and creating a ring of immune individuals around the outbreak by vaccination. Outbreaks lasted only for weeks, with case numbers rarely exceeding double figures; more than three-quarters of outbreaks ended with the generation that was infected immediately after the detection of infection.
R0 is a function of k,b and D; where k is the number of contacts each infectious individual has per unit time; b is the probability of transmission per contact between an infectious case and a susceptible person; and D is the mean duration of infectiousness4. In the case of infectious patients in hospital, their physical proximity to other patients and regular — often close and prolonged — contact with medical staff and nurses means that k and b will be greater than in the community or in domestic settings, particularly if transmission is only effective over short distances — centimetres rather than metres. This was the case for both SARS and smallpox, and nosocomial transmission was indeed a characteristic feature. In 45 outbreaks of variola major in Europe between 1950–1971, reviewed by Mack5, transmissions in hospital settings far outnumber those in any other category. Of the 680 cases, 339 (49.8%) contracted the disease in hospital, and 128 cases (18.8%) were in staff. In Singapore, 206 (76%) SARS cases were nosocomially acquired6, and of the 1,755 SARS infections in Hong Kong, 386 (22%) were in healthcare workers — 320 of whom were hospital staff who were infected while on duty7.
A low degree of transmissibility relative to other viruses is not the only property of the SARS virus that was an important factor in the success of the response to the outbreak. The ease of propagation and identification of SARS by well-established, standard virological methods were also of significance. The causative virus was grown without particular difficulty or problems with detection in readily available standard cell-culture systems. Within a month of the identification of SARS as a new clinical entity of uncertain aetiology — in late February 2003 in Hanoi, Vietnam — the virus had been grown in several laboratories. Researchers at the University of Hong Kong8 cultured the virus using foetal rhesus monkey kidney cells challenged with material from an open lung biopsy from a 53-year-old male Hong Kong resident, and a nasopharyngeal aspirate from a 42-year-old woman. Researchers at the Bernhard Nocht Institue for Tropical Medicine, Hamburg9 used Vero cells inoculated with sputum from a 32-year-old male physician who had fallen ill in New York, after travelling from Singapore where he had treated a patient. Finally, at the Centers for Disease Control and Prevention (CDC), Atlanta10, researchers cultured the virus in Vero cells inoculated with oropharyngeal samples from a 46-year-old male physician working in Vietnam, sputum from a 49-year-old male, a kidney sample from a 46-year-old male and oropharyngeal washings from an adult Vietnamese patient. In Hong Kong and Atlanta the results of thin-section and negative-staining electron microscopy showed unequivocally that the cytopathic effect was being caused by a coronavirus. In all three laboratories, reverse transcription PCR and comparison with known coronavirus sequences revealed the same sequence identity; this work also showed that the SARS virus was new, because its sequences were only distantly related to those from the wide range of previously sequenced coronaviruses that infect humans, dogs, cats, pigs, bovines, rats, mice, turkeys and chickens.
Influenza: lessons learned
Louis Pasteur said that “chance favours the prepared mind”. The relevance of this explanation of scientific success to the SARS virus, and in particular to the rapid initiation of collaborative work to understand and control it, is very clear. Influenza had prepared the way. Ever since the 1918–1919 influenza pandemic — which killed many millions, and was caused by a virus of subtype H1N1 — the possibility of the emergence of a virus strain capable of causing a similar event has, of necessity, been contemplated. The World Health Organization (WHO) initiated its international cooperation coordination programme in 1947. Pandemics with significantly lower mortalities occurred in 1957 (Asian flu, subtype H2N2), 1968 (Hong Kong flu, subtype H3N2) and 1977 (Russian flu, subtype H1N1) (Table 1). Although none have occurred since, the influenza outbreak that occurred in Hong Kong in 1997, although local and small in case numbers, had such a high mortality rate in confirmed cases that it stimulated a review of pandemic response policies both in Hong Kong and internationally. It meant that the Hong Kong Department of Health, Hospital Authority and laboratory surveillance facility11, and the WHO, were particularly well prepared to respond to the SARS outbreak.
Table 1.
Influenza landmarks in humans
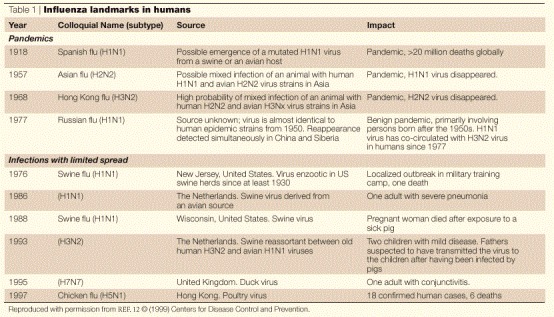
In March 1997, an outbreak of avian influenza caused by the A virus subtype H5N1 killed several thousand chickens in three rural Hong Kong chicken farms. In May 1997, a 3-year-old boy in Hong Kong contracted an influenza-like illness and died 12 days later from Reyes' syndrome — a paediatric complication that is associated with salicylate medication, which he had received. The virus strain resisted characterization with the available reagents; by August, detailed study in The Netherlands and the United States had revealed that the virus was closely related to the avian strains that were prevalent in March. In November, human cases caused by this virus began to occur; by late December, there had been 17 cases, of which five were fatal. Contact with chickens had occurred in all confirmed cases. On 28 December 1997, the slaughter of all chickens in Hong Kong (a total of 1.6 million) began, their import was stopped and the outbreak ceased.
In 1999, a group of influenza specialists reviewed the responses to the outbreak and drew several lessons from them12. Some were influenza-specific, but others had wider relevance. Regarding the actions that were taken in response to the outbreak, they reported that, at the time the outbreak started, the WHO was developing formal guidelines for addressing pandemic situations. These guidelines were revised after the Hong Kong influenza epidemic to include two strategic steps that were important in the outbreak — risk assessment (data collection and data evaluation) and risk management (continuously considering and reviewing the stages of a response, defining the risks and benefits, and making recommendations for the next steps to be followed). These principles underpinned the WHO response to SARS. The WHO Global Influenza Surveillance Network of four collaborating centres (in Atlanta, London, Melbourne and Tokyo) and 112 national influenza centres in 83 countries also had an extremely important role13. Reports in early February 2003 from Guandong Province, China, of 305 cases and five deaths owing to an atypical pneumonia of unknown aetiology, coupled with the isolation of an avian influenza virus A subtype H5N1 from two members of a family that had visited Fujian Province, China, in January, caused the activation of the pandemic plan. From analyses of samples taken from Vietnam, Singapore and Hong Kong, laboratories in the network ruled out the possibility of infection by any of the known influenza virus strains or other established causes of pneumonia, and concluded that SARS was new. On 15 March 2003, the WHO issued emergency travel advice and, using the influenza network as a model, set up a network of scientists from 11 laboratories around the world to identify the causative agent and develop diagnostic tests. Laboratories were chosen owing to their experience in detecting a wide range of microorganisms, a history of collaboration in international investigations coordinated by the WHO, their technical capacity to fulfil Koch's postulates and their access to SARS samples. Rules governing the confidentiality of data were set — shared data and information would only be used to advance the project in a collaborative way, data would only be shared outside the network with the approval of the originating laboratory, and samples would be regularly exchanged. There were daily teleconferences and a secure WHO website. On 16 April 2003, the participating laboratories collectively announced the conclusive identification of a new coronavirus as the causative agent. The WHO had performed well14.
Communication and microbiologists
Owing to its low R0 value and ease of cultivation, the challenge that SARS posed to the WHO was less difficult than it might have been. However, not everything was straightforward; for laboratories, epidemiologists and international organizations to react they must be told that there is a problem.
SARS. The reaction to SARS was delayed because initially it was unclear that there was a problem. The first cases of SARS probably occurred in Guangdong Province, China, in November 2002. On 23 January 2003, the health authority in Guangdong Province produced an expert report on the outbreak. Many of the conclusions that the health authority reached would have been of significant use to policy makers elsewhere, but circulation of this expert report was limited. Neither the WHO nor the Hong Kong authorities received a copy. Whether an early reversal of the denial of SARS by the Chinese authorities during the early part of the epidemic would have had a significant effect on later events can only be a matter for speculation. However, the WHO was not able to investigate the situation in China until April 12, by which time SARS had spread worldwide. A key factor in this spread had been the overnight stay in room 911 of the Metropole Hotel, Hong Kong, of a professor from Guangzhou in Guangdong, which triggered outbreaks in Hong Kong, Singapore, Canada and Vietnam. The lesson for microbiologists is a stark one — they operate in a political environment, and it can be unhelpful.
FMD. Other examples of the poor handling of infections by political systems are, unfortunately, not hard to find. There is a tradition in the United Kingdom of responding to disasters by holding rigorous inquiries afterwards, which are expected to leave the public feeling “confident that a searching investigation has been held, that nothing has been swept under the carpet and that no punches have been pulled”15. Although this is good, the pity is that the need for such inquiries seems to recur far too often. In 2001, the United Kingdom suffered a serious outbreak of foot and mouth disease (FMD). Millions of animals were slaughtered, and the overall costs totalled more than £8 billion. The conclusions of the 'Lessons to be Learned Inquiry'16 regarding the role of British virologists before and during the outbreak speak for themselves. The report states that, although the Pirbright Laboratory is the world reference laboratory and holds data on reported outbreaks throughout the world, “there is no coherent assessment of the full range of work undertaken by Pirbright in relation to the national surveillance and control strategies” and “the service arrangements in place in 2001 covered the processing of 300 samples a year...but contained no procedure for increasing the level of provision in an emergency”. In addition, “the Laboratory was not consulted when the FMD contingency plans were drawn up”.
BSE. The same failure to consult and use scientific expertise was evident in the handling of bovine spongiform encephalopathy (BSE) — politics came before science. Not long after the first occurrence of BSE, an attempt to describe a case in the scientific literature was firmly quashed by the Head of the Veterinary Investigation Service of England and Wales owing to “possible effects on exports and the political implications”17. The Phillips Inquiry into BSE and variant Creutzfeldt–Jacob Disease (CJD) reported the lessons that had been learned; the largest number of which addressed one topic — the improved use of scientific advisory committees. Their failure to use the full range of expertise available — even within the United Kingdom — contrasts sharply with the inclusive approach adopted by the WHO in the case of the SARS epidemic.
Microbiology and the public
The United Kingdom was lucky with SARS as there were no reported outbreaks. However, the situation was very different in Hong Kong, which had 1,755 cases and 300 deaths. An expert review committee appointed by the Government of the Hong Kong Special Administrative Region reported its findings in October 2003 (Ref. 7). Among other things, the committee was charged with the identification of lessons to be learned and with making recommendations for future epidemics. They focused on seven principles: a strengthened epidemiology capacity, systems for early detection and reporting, contingency planning, clear command and control structures, integrated responses, a surge capacity and transparency and effective communication. Although it is impossible to disagree with these principles and their importance in outbreak control, a striking feature of the 279-page report is that, with the exception of a box describing the isolation of the virus in Hong Kong entitled 'Ground-breaking discovery' early in the report, there is little comment, favourable or unfavourable, about virology, virology laboratories or virologists. It could be said that this might be a reflection of the backgrounds and interests of the committee that generated the report — the two co-chairs (both from the United Kingdom) were experts in hospital management and administration and in public health, respectively. But it is very probable that it reflects another problem — the lack of regard that is paid to the importance of microbiology in the response to microbiological threats to public health. Reforming and improving health-service administrative structures and information systems, and planning for unexpected rises in the number of contagious patients are all necessary, but by themselves they are insufficient. The flaw in Hegel's aphorism about lessons from history is that microorganisms evolve in real time; learning lessons from past failures (and successes) will be insufficient because evolution throws up new challenges. In their review18, MacLehose, McKee and Weinberg state that “one of the greatest challenges facing surveillance systems is awareness of the unexpected, recognizing when things seem not quite right. Nipah virus was thought to be Japanese B encephalitis, West Nile virus in New York was thought to be St Louis encephalitis, and prions were thought not to cross species barriers. Focusing surveillance systems on the diseases of today fails to address the challenges of an uncertain future”.
We were lucky with SARS due to the existence of a network of laboratories that were looking for new influenza viruses and which were linked by the mature surveillance network run by the WHO. Other microbiological networks also exist. For food-borne pathogens there are, for example, Pulse Net19 in the United States (which uses standardized pulsed-field gel electrophoresis (PFGE) protocols to generate and compare profiles of Escherichia coli O157, Salmonella spp. and Listeria monocytogenes), and Enternet (which uses phenotypic data for typing), Salmgene and PulseNet Europe (both of which use PFGE) in Europe. The European Commission has proposed the creation of a European Centre for Disease Control and Prevention20 to standardize surveillance methods, ensure data compatability, provide scientific assessments, technical support, information to officials and to the public, and to assume responsibility for existing networks and early warning and response systems. However, it is proposed that it will only have a staff of 15. The European Health Commissioner David Byrne explained that “it will be a hub, an intelligence centre. The core is already in place”. In this way it will be utterly reliant on national laboratories, national microbiologists and national infrastructures. Is this 'core' up to the job? Consider the United Kingdom. Its core is at least as effective as that of any other large European country. In universities, for example, it has many microbiologists of international rank21. However, recent inquiries have identified serious problems. The recent report of the House of Lords Select Committee on Science and Technology, 'Fighting infection'22, concluded that the infectious disease services in England that are “expected to protect the population from both common and more unusual infections are under-resourced and over-stretched...there is not enough surge capacity”. The report went on to predict that “without improvements we fear that this country will suffer from major epidemics”. The Academy of Medical Sciences inquiry23 into 'Academic medical bacteriology in the twenty-first century' focused on research. The inquiry concluded that although bacteriology has recently undergone a transformation with the introduction of several new techniques, “medical microbiology departments in the United Kingdom, with a few exceptions, are in a state of torpor...”. So microbiologists need to get their act together, and it will not do to rely on the bursts of funding and political approval that result from the panic engendered by disease outbreaks. Hegel was right; history tells us that this kind of support is short-lived. SARS proves the point. In July 2003, the WHO declared the world to be free of SARS, and in August 2003, Klaus Stöhr, its SARS research coordinator, stepped down and returned to influenza work owing to a lack of money24.
Concluding remarks
The ability to respond rapidly to new pathogens is not cheap. The next pathogen might have a high R0 value and be as elusive as a prion. The past tells us to be prepared. European budgets pale into insignificance compared with the $6.5 billion of the US CDC. Obviously, European microbiologists have lessons to learn about getting political support! But funding is not the whole answer. The United Kingdom, for example, has plenty of microbiological talent. Recent experiences with BSE and FMD indicate the need for big improvements in how policy makers get scientific advice. This problem persists. Doctors, veterinarians, civil servants, politicians and scientists must come out of their boxes, learn to speak their different languages and communicate more effectively. If they fail, sooner or later there will be a heavy price to pay. Evolution says so.
Biography
Hugh Pennington is Emeritus Professor of Bacteriology at Aberdeen University and President of the Society for General Microbiology. He chaired an inquiry for the UK Government into the 1996 Escherichia coli O157 outbreak in central Scotland, and, at present, is a member of committees that advise the UK Food Standards Agency, the World Food Programme and the BBC.
Related links
FURTHER INFORMATION
CDC information on selected threats and diseases
Competing interests
The author declares no competing financial interests.
References
- 1.Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 2.Anderson RM, May RM. Infectious Diseases of Humans. 1991. [Google Scholar]
- 3.Gani R, Leach S. Transmission potential of smallpox in contemporary populations. Nature. 2001;414:748–751. doi: 10.1038/414748a. [DOI] [PubMed] [Google Scholar]
- 4.Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mack TM. Smallpox in Europe, 1950–1971. J. Infect. Dis. 1972;125:161–169. doi: 10.1093/infdis/125.2.161. [DOI] [PubMed] [Google Scholar]
- 6.Singh K, et al. Severe acute respiratory syndrome: lessons from Singapore. Emerg. Infect. Dis. 2003;9:1294–1298. doi: 10.3201/eid0910.030388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SARS Expert Committee. SARS in Hong Kong: from experience to action [online], (cited 16 Feb 2004), <http://www.sars-expertcom.gov.hk/english/reports/reports.html> (2003).
- 8.Peiris JSM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten C, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Eng. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Eng. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.Normile D. Up close and personal with SARS. Science. 2003;300:886–887. doi: 10.1126/science.300.5621.886. [DOI] [PubMed] [Google Scholar]
- 12.Snacken R, Kendal AP, Haaheim LR, Word JM. The next influenza pandemic: lessons from Hong Kong, 1997. Emerg. Infect. Dis. 1999;5:195–203. doi: 10.3201/eid0502.990202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization multicentre collaborative network for severe acute respiratory syndrome diagnosis. A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet361, 1730–1733 (2003). [DOI] [PMC free article] [PubMed]
- 14.Report of the UK Academy of Medical Sciences working group on SARS. [online], (cited 16 Feb 2004), <http://www.acmedsci.ac.uk/n_sars_grp.htm> (9 June 2003).
- 15.UK Department of Transport. MV Herald of Free Enterprise Report of Court No. 8074. The Merchant Shipping Act Formal Investigations 1894. (The Stationery Office, London, 1987).
- 16.Anderson, I. Foot and mouth disease 2001. Report of the 'Lessons to be learned' inquiry Report. [online], (cited 16 Feb 2004), <http://213.121.214.218/fmd/report/index.htm> (2002).
- 17.Pennington TH. When Food Kills. 2003. [Google Scholar]
- 18.MacLehose L, McKee M, Weinberg J. Responding to the challenge of communicable disease in Europe. Science. 2002;295:2047–2050. doi: 10.1126/science.1070025. [DOI] [PubMed] [Google Scholar]
- 19.The National Molecular Subtyping Network for Foodborne Disease surveillance. [online], (cited 16 Feb 2004), <http://www.cdc.gov/pulsenet> (2003).
- 20.Bosche X. European equivalent of US Centers for Disease Control proposed. Lancet. 2003;362:455. doi: 10.1016/S0140-6736(03)14103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.2001 Research Assessment Exercise: The Outcome [online], (cited 16 Feb 2004), <http://195.194.167.103/Pubs/4_01/> (2001).
- 22.House of Lords Select Committee on Science and Technology Session 2002–2003 Fourth Report. Fighting Infection. [online], (cited 16 Feb 2004), <http://www.parliament.the-stationery-office.co.uk/pa/ld200203/ldselect/ldsctech/138/13803.htm> (2003).
- 23.UK Academy of Medical Sciences. Academic Medical Bacteriology in the twenty-first century [online], (cited 16 Feb 2004), <http://www.acmedsci.ac.uk/ac_bac.pdf> (2001).
- 24.Enserink M, Normile D. Search for SARS origins stalls. Science. 2003;302:766–767. doi: 10.1126/science.302.5646.766. [DOI] [PubMed] [Google Scholar]


