Figure 4. Positive selection of bat genes.
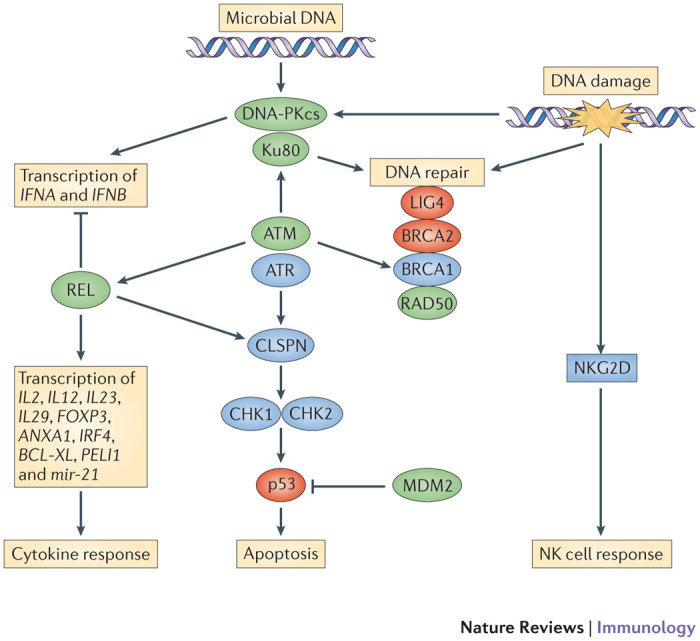
The figure illustrates key components of the DNA damage response and DNA repair pathways. Whole-genome analysis of two bat species (Pteropus alecto and Myotis davidii) showed that a high number of genes encoding components of these pathways are positively selected in P. alecto and M. davidii. Many of these genes are positively selected in both species (these encode proteins that are highlighted in green), whereas others have been positively selected in only one of the species (these encode proteins that are highlighted in red). Bat-specific differences in these genes include unique changes to the nuclear localization signals in p53 and the E3 ubiquitin-protein ligase MDM2. The impact of these changes on host–pathogen interactions is currently under investigation. The transcription factor REL (which is a member of the NF-κB family) regulates many effector proteins of the innate immune system, including type I interferons (IFNs), and it also participates in the DNA damage response. The gene encoding REL is under positive selection in bats and contains unique amino acid changes that might affect the interaction of REL with inhibitor of NF-κB (IκB). DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Ku80 are crucial to the DNA damage response but also form a microbial DNA-sensing complex connected to the type I IFN system. The relationship between innate immunity and the DNA damage response is well established, and unique changes in this pathway in bats might influence the outcomes of viral infection. It was proposed that these differences might reflect adaptations to the increased oxidative metabolism that accompanied the evolution of flight in bats. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related protein; CHK, checkpoint kinase; CLSPN, claspin; FOXP3, forkhead box P3; IL, interleukin; LIG4, DNA ligase 4; MRE11, meiotic recombination 11 homologue; NBS1, Nijmegen breakage syndrome protein 1 (also known as nibrin).
