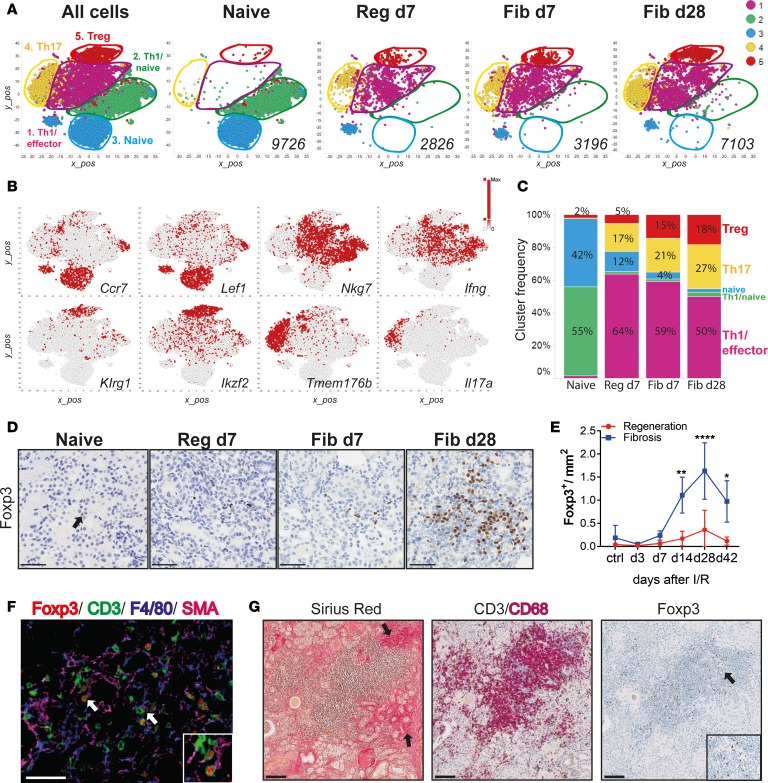
Figure 3. Single-cell RNA-Seq of kidney CD4+ T cells identifies a population of regulatory T cells that expands during fibrosis.
(A) Unsupervised cell clustering colored by the 5 major populations from sorted kidney CD4+ T cells in 2-dimensional t-SNE plots. A total of 22,851 combined kidney single cells (left) and separated by indicated conditions and time points (right). Numbers indicate the total number of cells per condition. (B) t-SNE plots showing selected marker genes for each cluster. Colors indicate the gene expression level. (C) Bar graphs showing the frequency of each cluster (as in A) per condition and time point. (D) Representative Foxp3 immunostaining on kidney sections from naive, regeneration day 7, and fibrosis days 7 and 28, and (E) morphometric quantification. (F) Multicolor immunofluorescence of kidney sections from fibrosis day 28. Arrows indicate Tregs interacting with SMA+ (zoomed area in the square insert at the bottom right) and F4/80+ cells. (G) Kidney biopsies from a patient after transplant with fibrosis, stained with Sirius Red and for CD3/CD68 and Foxp3. Arrows on the left picture indicate fibrotic areas and the right picture shows Treg accumulation (zoomed areas in the small square). Scale bars: 50 μm (D and F) and 100 μm (G). Results are representative of 1 (regeneration model) or 2 (fibrosis model) independent experiments, with 3 to 5 mice per time point/condition. Mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001 compared with controls by 2-way ANOVA followed by Dunnett’s post hoc test for multiple comparisons (E). Reg, regeneration; Fib, fibrosis.