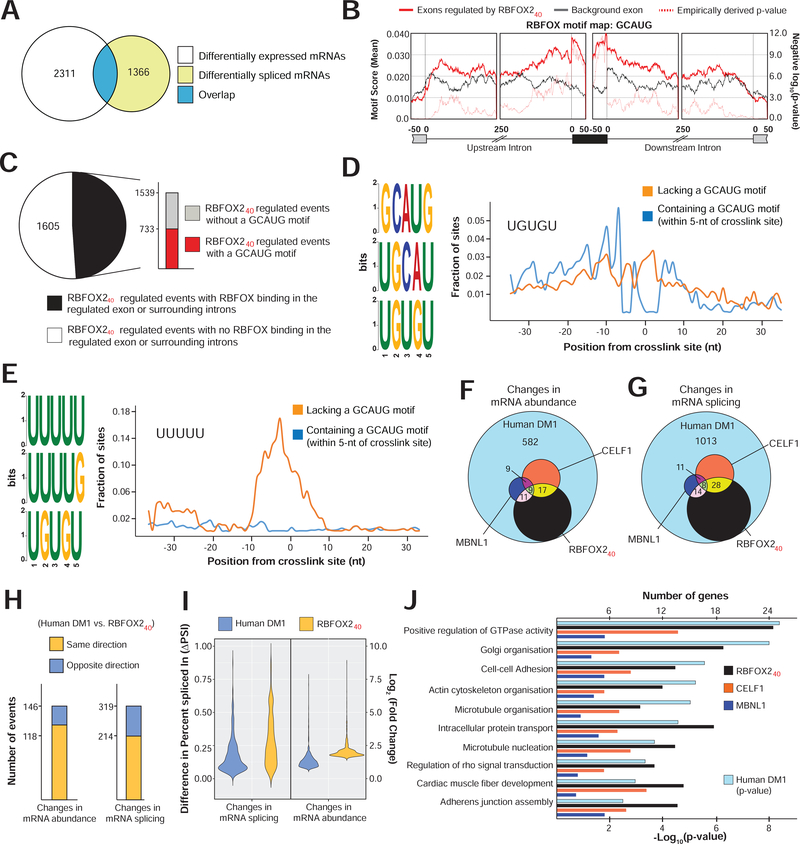
Figure 5. RBFOX240 isoform driven transcriptome alterations in DM1 heart tissue.
(A) Overlap of differentially expressed (p<0.05, Wald test as described DESeq2; Log2[Fold Change]>1, and TPM>4) with differentially spliced mRNAs (p<0.05, FDR<0.10 adjusted for multiple testing, Difference in Percent Spliced Index [ΔPSI]≥20%, and Junction Counts≥10) in cardiomyocytes isolated from hemizygous MHCrtTA, and TRE-RBFOX240; MHCrtTA bitransgenic mice after administration of 0.5g/kg Dox containing Chow for 3 days. (B) Position and relative enrichment of RBFOX-binding motif near RBFOX240 regulated cassette exons. (C) Breakup of 3144 RBFOX240 regulated splicing events in cardiomyocytes with or without RBFOX240 binding peaks and a GCAUG motif within those peaks from iCLIP data in mouse brain (Damianov et al., 2016). (D-E) Top 3 pentamers enriched within the binding peaks near RBFOX240 regulated exons with (D) or without (E) GCAUG motifs. Fractional enrichments of (D) UGUGU and (E) UUUUU sequences aligning at each nucleotide relative to the RBFOX240 crosslink site are also plotted. Overlap of (G) mRNA abundance, and (F) alternative splicing changes among cardiac transcriptomes of DM1 patients (Freyermuth et al., 2016), RBFOX240 overexpressing, CELF1 overexpressing, and Mbnl1ΔE3/ΔE3 mice (Wang et al., 2015). (H) Directionality of mRNA abundance and alternative splicing changes in DM1 patient hearts and RBFOX240 overexpressing cardiomyocytes. (I) Distribution of changes in mRNA abundance and splicing events co-regulated in DM1 patient hearts and RBFOX240 overexpressing cardiomyocytes. (J) p-values of top Gene Ontology terms for alternatively spliced mRNAs in DM1 patient hearts (hypergeometric test; Benjamini method for multiple testing), and corresponding numbers of genes for each category that are similarly misspliced in the cardiomyocytes of RBFOX240 overexpressing, and hearts of CELF1 overexpressing, or Mbnl1ΔE3/ΔE3 mice.