Fig. 1.
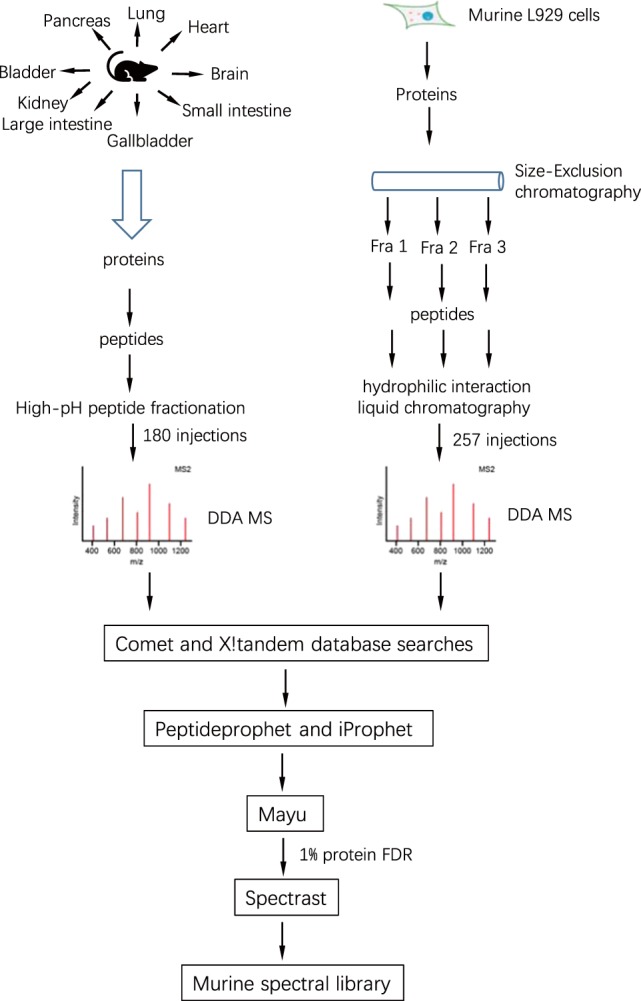
Sample preparation and data analysis workflows used in the generation of the spectral library. L929 cell lysates were first fractionated with size-exclusion chromatography and digested with trypsin. The resulting peptides were fractionated with HILIC (Hydrophilic Interaction Liquid Chromatography). The tissue samples were digested with trypsin and the peptides were fractionated with high-pH chromatography. The peptide fractions were dissolved in 0.1% formic acid containing iRT peptides, which were analyzed using shotgun MS. The DDA files were searched with X!Tandem and Comet, and results were combined with iProphet. The combined results were filtered with 1% protein FDR and made a consensus spectral library with Spectrast software.
