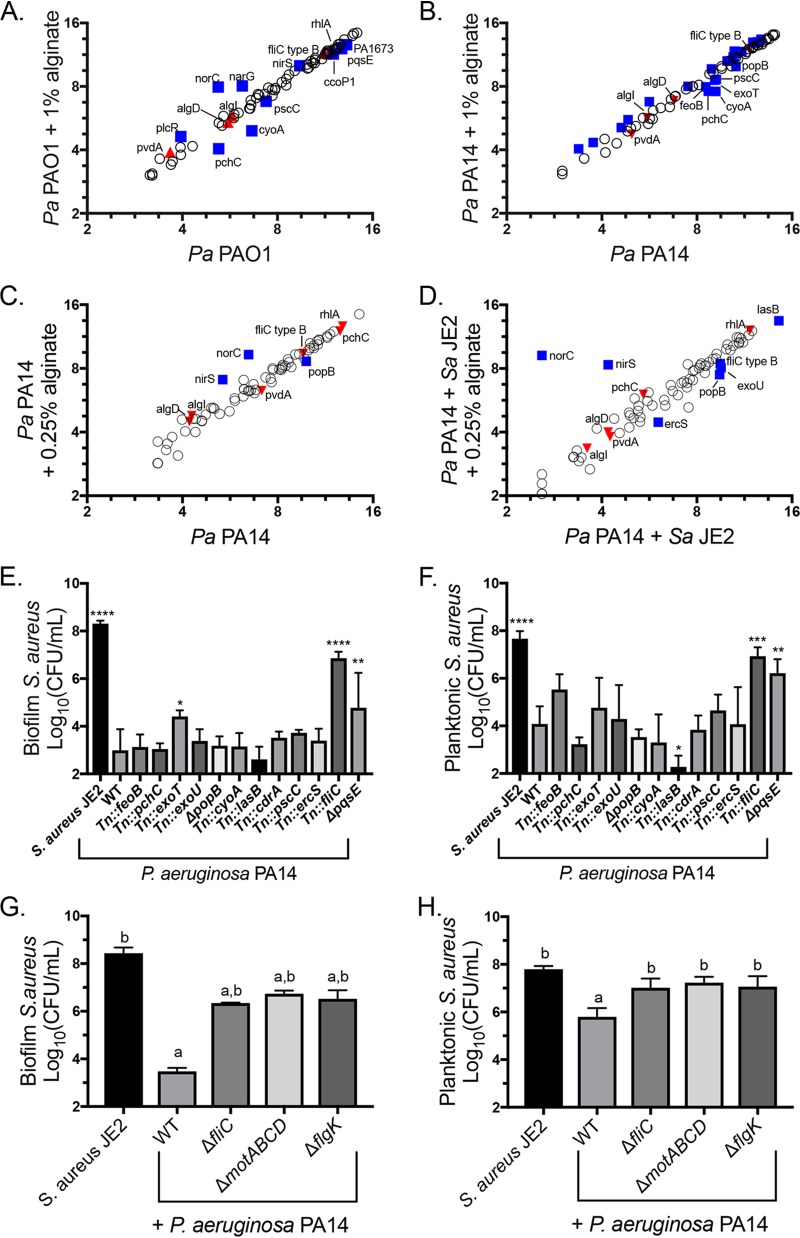
FIG 7.
Alginate alters the expression of P. aeruginosa genes essential for S. aureus killing. (A to D) Raw Nanostring counts were normalized to values for the positive controls and three housekeeping genes (rpoD, ppiD, and fbp) and log2 transformed. Three biological replicates were performed under each condition. Significantly differentially expressed genes were determined by an unpaired t test followed by the two-stage linear step-up procedure of Benjamini et al. (57) (with a q value of 1% for false discovery) and are marked by blue squares and labeled with the gene name. Genes significantly differentially expressed by mucoid P. aeruginosa in a previous study (32) but not by P. aeruginosa in the presence of exogenous alginate are marked by red triangles. (A and B) P. aeruginosa PAO1 (A) and P. aeruginosa PA14 (B) subcultured into TSB or TSB plus 1% alginate during the mid-log growth phase for 45 min. For clarity, only significantly downregulated genes are labeled in panel B. (C and D) P. aeruginosa PA14 monocultured with 0.25% alginate for 8 h (C) and cocultured with S. aureus JE2 in TSB with 0.25% alginate for 8 h (D). (E and F) S. aureus JE2 survival in the biofilm (E) and planktonic (F) fractions after 16 h of coculture with the indicated P. aeruginosa PA14 mutants corresponding to genes downregulated in one or more Nanostring experiments. Significance was determined by one-way ANOVA with Dunnett’s posttest comparison to S. aureus plus P. aeruginosa PA14. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G and H) S. aureus JE2 survival in the biofilm (G) and planktonic (H) fractions after 16 h of coculture with P. aeruginosa PA14 motility mutants. Significance was determined by one-way ANOVA with Dunnett’s posttest. a, P < 0.05 with S. aureus JE2 as the reference; b, P < 0.05 with P. aeruginosa PAO1 plus S. aureus JE2 as the reference. WT, wild type.