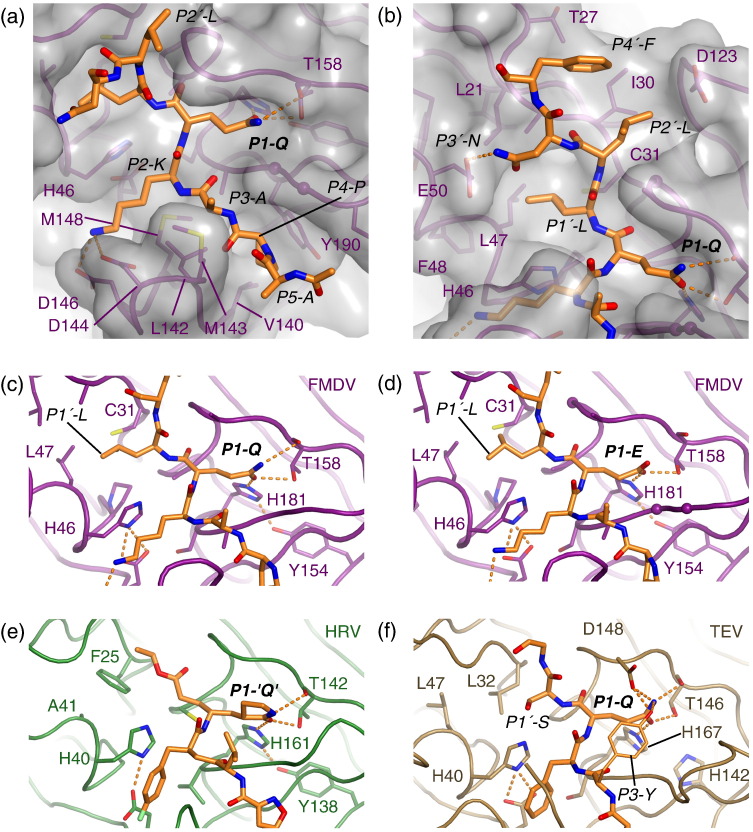
Fig. 2.
Structural details of peptide recognition by FMDV 3Cpro. (a) Close-up view of the binding of the P5–P1 residues from the VP1-2A peptide. The protease backbone is depicted as a smooth ribbon covered by a semitransparent gray van der Waals surface; selected side chains are shown. (b) Close-up view of the binding of the P1′–P4′ residues from the VP1-2A peptide. Detailed views of the P1–P1′ interactions for (c) FMDV 3Cpro complexed with VP1-2A and for (d) FMDV 3Cpro complexed with VP1-2Am. The two hydrogen bonds made by OE1 from the P1-Glu side chain to Thr158 and His181 are shown as dotted lines; atom OE2 from the P1-Glu side chain is too distant from Thr158 to form a hydrogen bond. (e) HRV 3Cpro complexed irreversibly with a peptide-like inhibitor.25 (f) Tobacco etch virus NIa 3C-like protease complexed with a P7–P3′ peptide.28