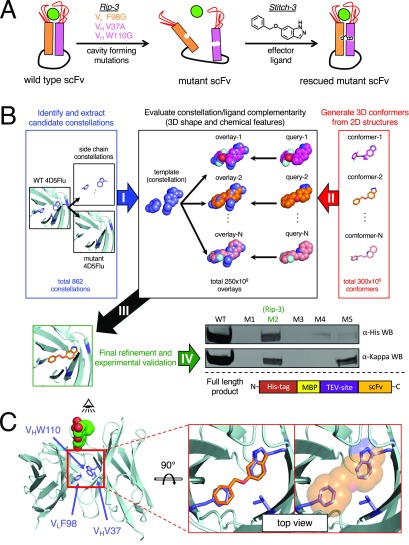
Figure 1.
Design of switchable antibodies. (A) Summary of the design strategy. Introducing cavity-forming mutations in the interface between the heavy and light chains will lead to dissociation of this domain–domain interface, leading to loss of antigen-binding activity. Subsequent addition of a rescuing ligand will induce reassociation of the domain–domain interface and, thus, restore activity. (B) Computational design strategy. (I) All possible combinations of two/three-residue cavity-forming mutations at this domain/domain interface are exhaustively considered, to identify those yielding a suitable cavity for subsequent rescue by a druglike small molecule. (II) Energetically favorable three-dimensional conformations (“conformers”) are generated from each member of a large compound library. (III) For each “constellation of atoms” that can be deleted from the protein domain/domain interface, potential structural matches are identified from the library of three-dimensional compound conformations. (IV) The top-scoring structural matches are refined in the context of the (mutant) protein environment, and the best 5 resulting designs are selected for experimental characterization. Using anti-His and anti-kappa (light chain) Western blots, we find that only M2 is solubly expressed in its complete form (the uncropped Western blots are shown in Figure S1); we therefore focused further characterization on this design, which we refer to as Rip-3/Stitch-3. (C) Design model of the rescued Rip-3/Stitch-3 complex. The crystal structure of 4D5 (which harbors the same framework as 4D5Flu) was used as a starting point, and the fluorescein antigen was modeled from a separate antibody (4-4-20). The residues that comprise the Rip-3 constellation are indicated in blue sticks and are shown in superposition with the ligand predicted to rescue this mutation (Stitch-3, orange).