Figure 3:
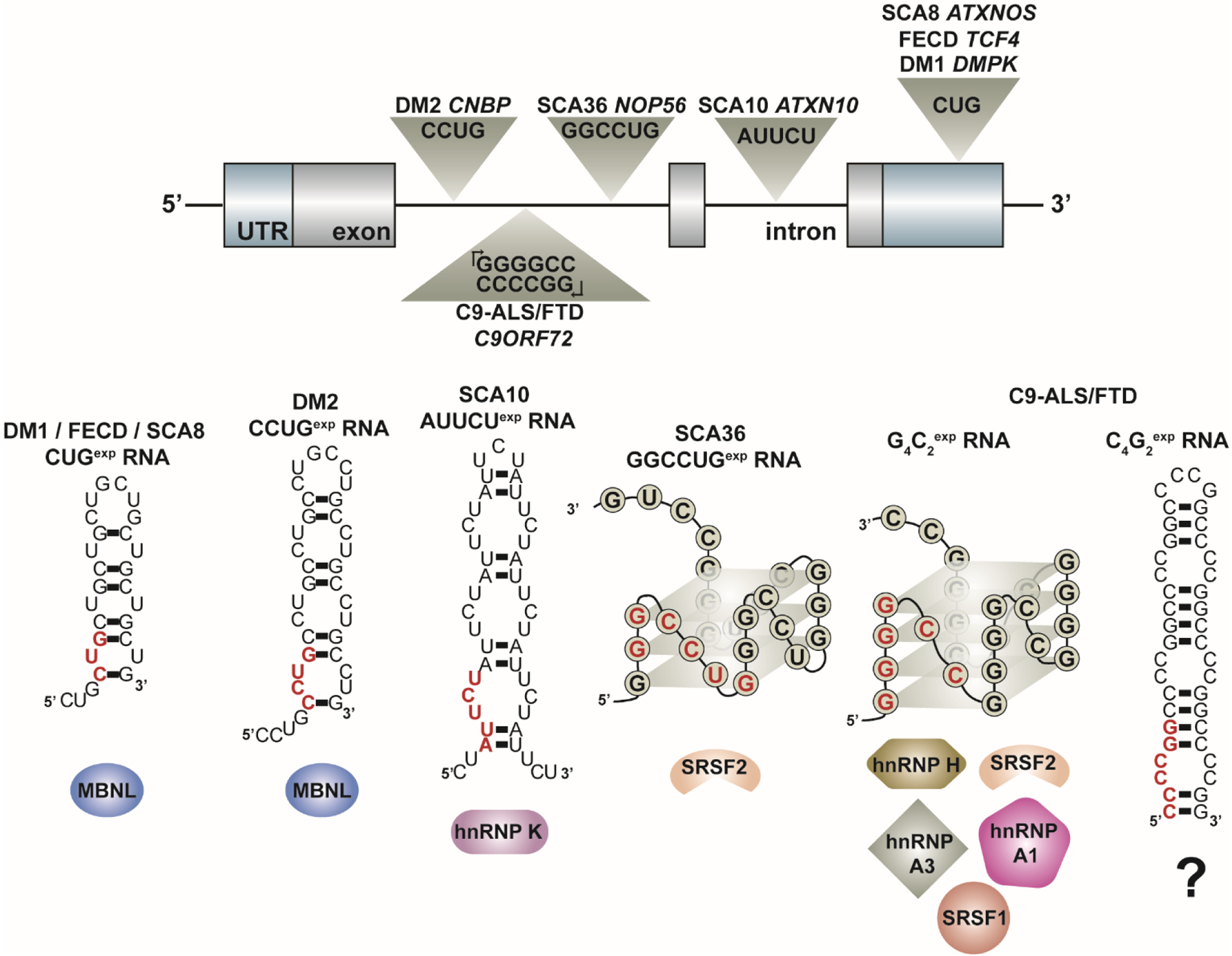
Expanded repeat RNAs transcribed from many microsatellite repeat expansions associated with genetic disease form secondary structures in vitro predicted to sequester RBPs that regulate alternative splicing. These expanded repeats are located within different regions of genomic architecture and can be transcribed bi-directionally (see C9-ALS/FTD). In vitro studies indicate that these repeat RNAs form secondary structures that classify into two general groups – A-form RNA helices or G-quadruplexes – as shown by these model RNA structures. A single repeat unit is highlighted in red in each model RNA structure. These toxic RNAs are found within ribonuclear foci and have been shown to co-localize with several RBPs that act as alternative splicing (AS) factors as listed below each RNA structure. The sequestration of these RBPs leads to disease-associated mis-splicing of multiple target pre-mRNAs. Outstanding questions in the field include (1) what structure(s) these repeat RNAs adopt in vivo and (2) the role of RNA secondary structure in modulating the interaction of RBPs with the toxic RNA and subsequent downstream dysregulation of alternative splicing and other RNA metabolic processes.
