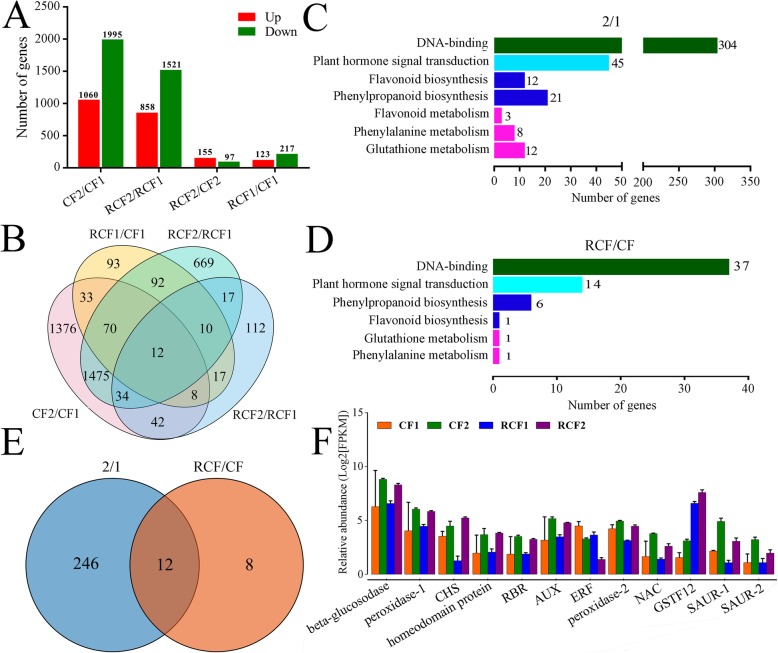
Fig. 2.
Number of differentially expressed genes (DEGs) identified by RNA-seq analysis. a Numbers of DEGs. b Venn diagram representing numbers of DEGs. c Number of DEGs in group 2–1 categorized into DNA binding, plant hormone signal transduction, flavonoid biosynthesis, phenylpropanoid biosynthesis, brassinosteroid biosynthesis, zeatin biosynthesis, flavonoid metabolism, phenylalanine metabolism, glutathione metabolism and ABC transporters. d Number of DEGs in group RCF-CF involved in DNA binding, plant hormone signal transduction, phenylpropanoid biosynthesis, flavonoid biosynthesis, glutathione metabolism and phenylalanine metabolism. e Overlapping DEGs between group 2–1 and group RCF-CF. f Transcript levels of overlapping DEGs between group 2–1 and RCF-CF as determined by RNA-seq