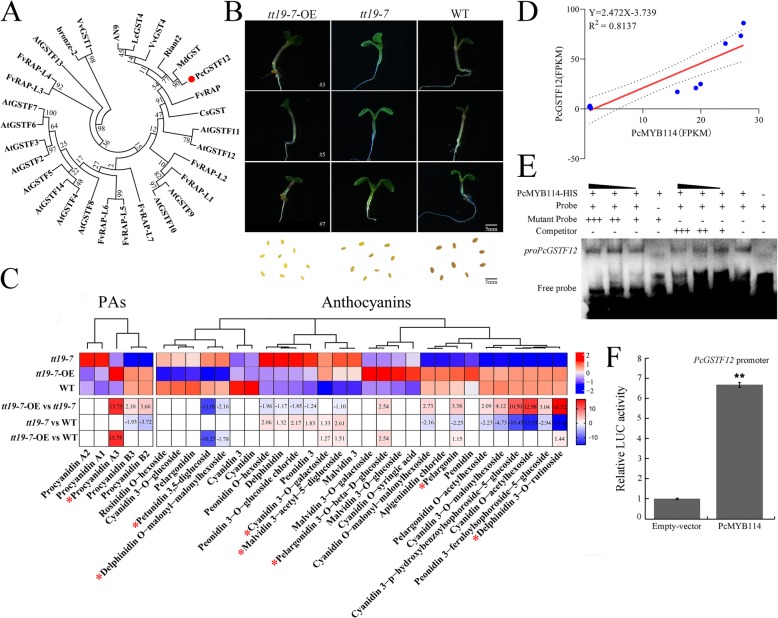
Fig. 5.
Functional analysis of PcGSTF12 in anthocyanin and procyanidin (PA) accumulation. a Phylogenetic analyses of PcGSTF12 and its paralogs. The accession numbers are MdGST (AEN84869), Riant2 (KT312848), LcGST4 (KT946768), AN9 (Y07721), FvRAP-L1 (gene28763), VvGST4 (AAX81329), CsGST (ABA42223), AtGSTF2 (At4G02520), AtGSTF5 (At1G02940), AtGSTF3 (At2G02930), bronze-2 (AAV64226), FvRAP-L7 (gene10552), AtGSTF12/TT19 (At5G17220), AtGSTF14 (At1G49860), AtGSTF7 (At1G02920), VvGST1 (AAN85826), FvRAP (gene31672), FvRAP-L4 (gene10549), FvRAP-L2 (gene08595), FvRAP-L3 (gene22014), AtGSTF10 (At2G30870), FvRAP-L5 (gene10550), AtGSTF11 (At3G03190), AtGSTF13 (At3G62760), FvRAP-L6 (gene10551), AtGSTF8 (At2G47730), AtGSTF4 (At1G02950), AtGSTF6 (At1G02930), AtGSTF9 (At2G30860). b Phenotypes of tt19–7-OE seedlings and fresh seeds. c Heat map of anthocyanins and PAs of 7-day-old seedlings of tt19–7 mutant, tt19–7-OE, and WT (wild-type). tt19–7-OE refers to 35S::PcGSTF12 transgenic lines. Scale bars: 5 mm (B–C). Differentially accumulated metabolites between WT and tt19–7-OE marked by red star. d Correlation analyses between transcript levels of PcGSTF12 and PcMYB114. e EMSA assays. Probe was biotin-labeled fragment containing MBS motif. Competitor probe was non-labeled probe. Mutant probe contained two nucleotide mutations. Competitors, and mutant probes at a 10×, 25×, and 50× molar excess were present (+) or absent (−) in each reaction. Black arrows indicate increasing multiples. f Effects of PcMYB114 on promoter activities as demonstrated by luciferase reporter assays. Empty-vector was used as a control. Values are means ± SD of three independent biological replicates. Statistical significance: **P < 0.01