Figure 3. Design of new 5′ UTRs.
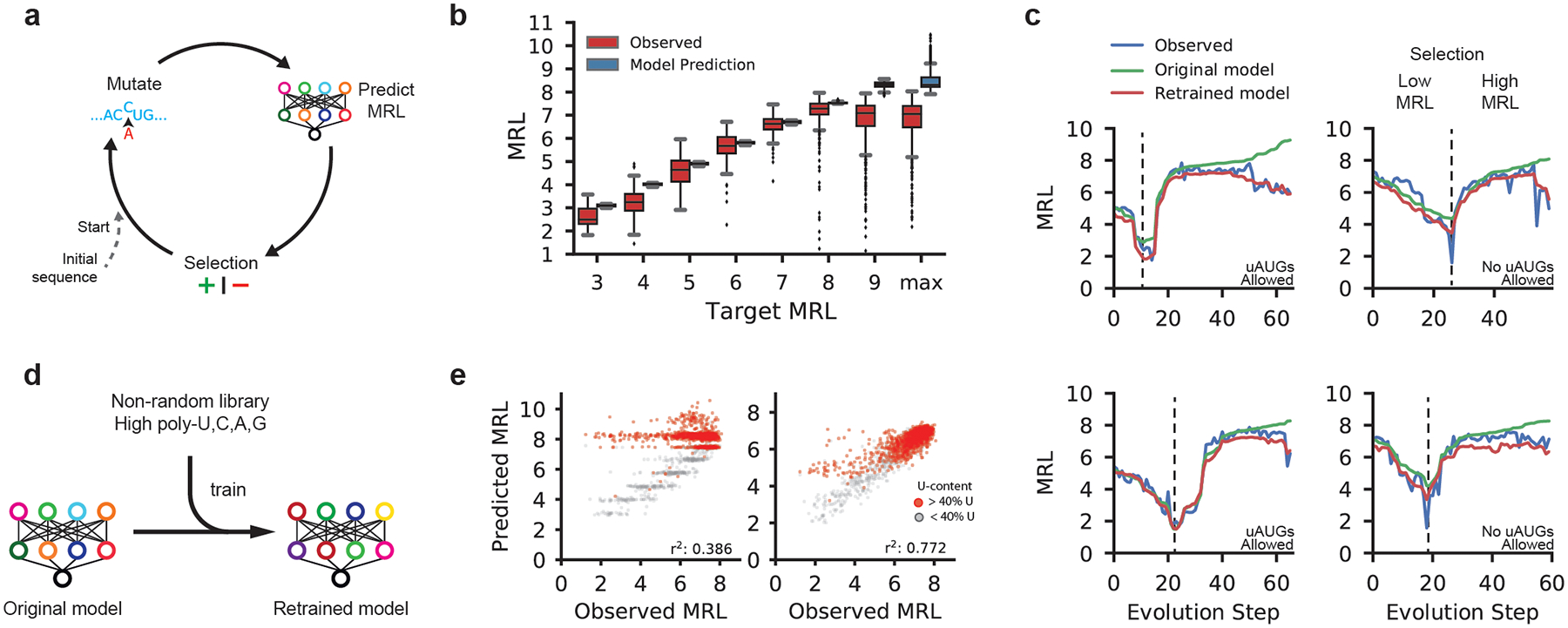
(a) Diagram of a genetic algorithm that was used in conjunction with Optimus 5-Prime to evolve sequences to target specific levels of ribosome loading. (b) Comparison between the predicted MRLs and observed MRLs for evolved 5′ UTRs for targeted ribosome loading. All 16 box plots are defined in terms of the sample size, minima, median, maxima and percentiles (Supplementary Table 3). (c) Step-wise evolution analysis. Randomly initialized UTRs were first evolved for low ribosome loading and then for high ribosome (selection change at dashed line). Four out of 80 (Supplementary Fig. 11a–d) examples are shown. Examples on the left were permitted to have uAUGs while those on the right were not. Each unique sequence that was generated during the evolution process was synthesized and tested by polysome profiling. The original Optimus 5-Prime prediction (green) and the observed MRL eventually diverge, but the predictions from the retrained Optimus 5-Prime (red) more accurately reflect the data. (d) The original Optimus 5-Prime is retrained using sequences from the designed library with high poly-U, C, A, and G stretches which occur rarely in the random library. (e) The accuracy of the retrained Optimus 5-Prime increased when predicting the high poly-U sequences (red) generated by the genetic algorithm (r2: 0.386 to 0.772, n = 2,146).
