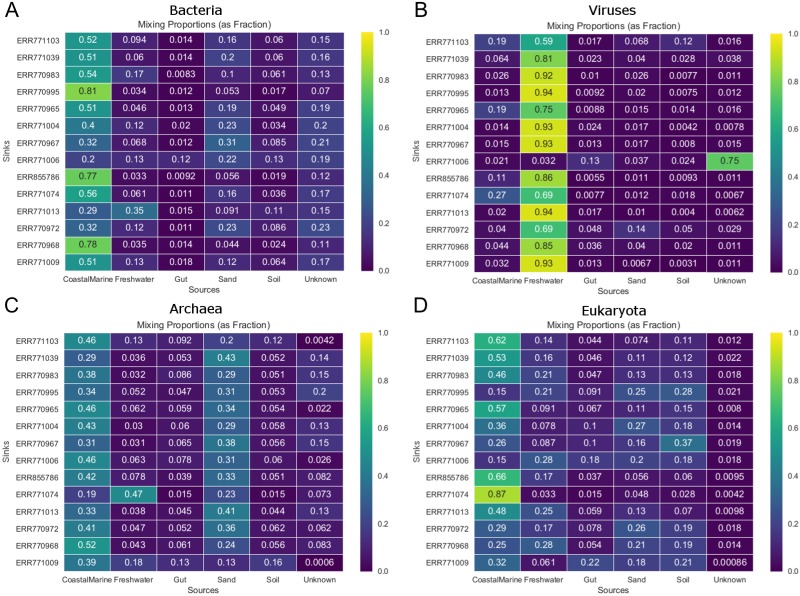
Figure 3. Taxon-dependent source proportion estimates for 14 different coastal marine samples.
Metagenome data was separated into taxa groups (see Methods) and multiple coastal marine samples were designated as “sinks”. Heatmaps produced by mSourceTracker represent the proportions of (A) Bacteria, (B) Viruses, (C) Archaea, and (D) Eukaroyta from each of the source environments. mSourceTracker default number of chains was changed to 5, and number of draws were adjusted per taxa group so absolute values between any 2 Markov chains did not exceed 5%. Five chains were used for every environment, but the number of draws was changed depending on the domain (100 draws for Archaea, 80 draws for Eukaryota, 50 draws for viruses, 20 draws for Bacteria, and 20 draws for the combined dataset).