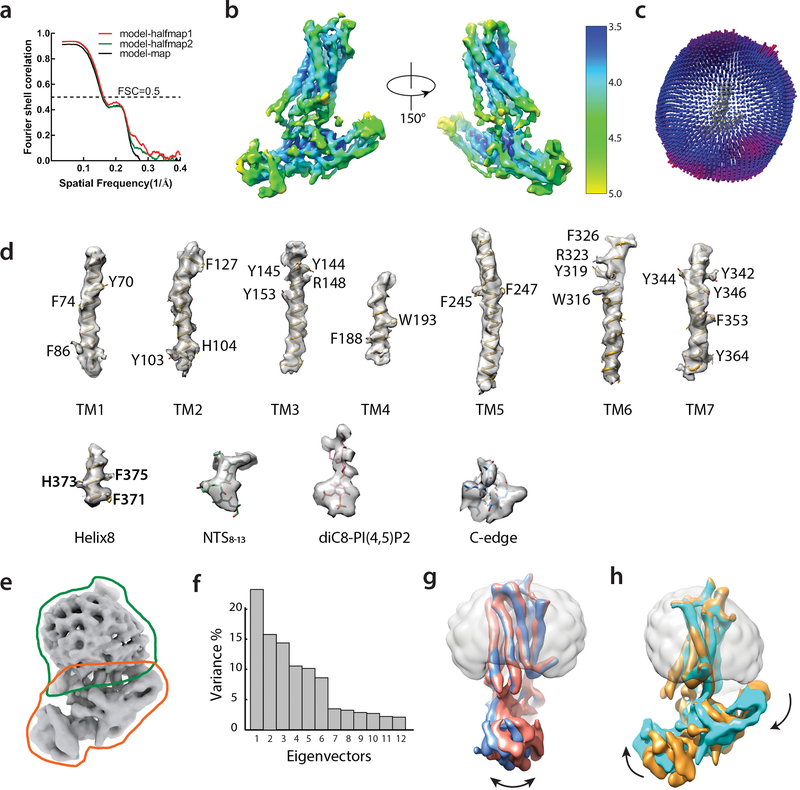
Extended Data Figure 6: CryoEM map resolution and model validation, with multi-body analysis revealing structural heterogeneity within the NTSR1-βarr1ΔCT complex.
a) Map-model cross validation. The model was refined against one half map after displacement of atoms by 0.2 Å, and FSC curves were calculated between this model and the working half1 map (red), the free half2 map (green) and the final cryoEM map (full dataset, black) by Mtriage implemented in Phenix. b) Local resolution of the final 4.2 Å map was estimated by Bsoft. c) Euler angle distribution of particle set used in the final map. d) Representative sections of the model with accompanying regions of density from the EM map. e) Masks employed for multi-body refinement. f) The contributions of each of the twelve eigenvectors (numbered along x-axis) to the variance of the overall final map. g) Maps corresponding to the 2nd and 9th components of the first eigenvector (panel f) are aligned, showing swing-like motion of βarr1ΔCT with respect to NTSR1. h) Maps corresponding to 1st and 10th components of the second eigenvector (panel f) are superimposed, indicating tilt-like motion of βarr1ΔCT with respect to NTSR1.