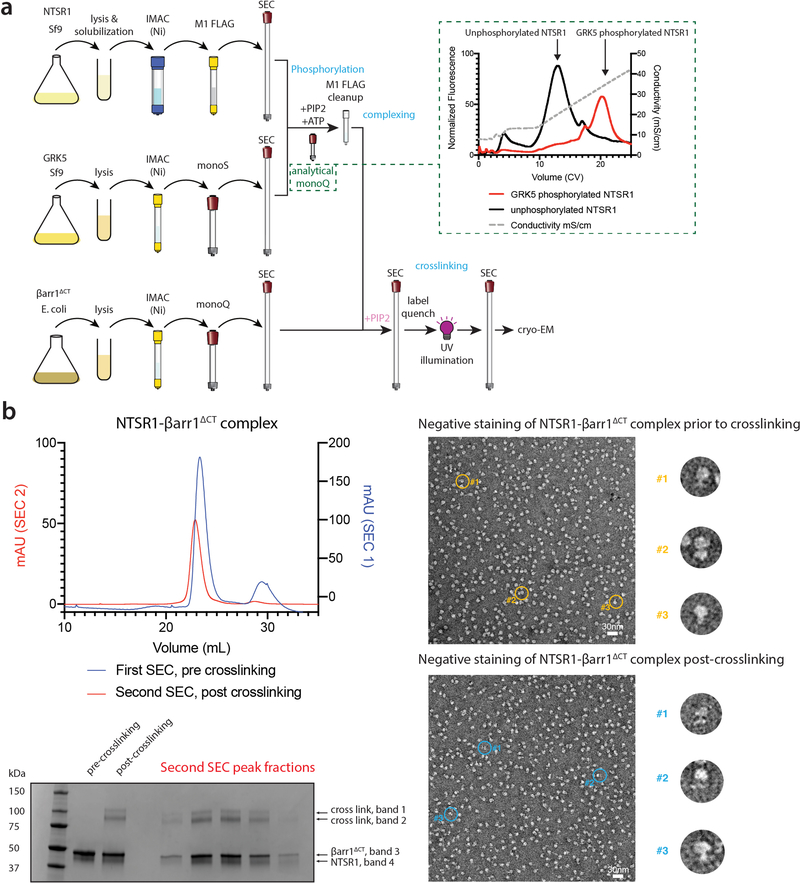
Extended Data Figure 3: Overview of sample preparation and representative cryoEM preparation.
a) Workflow for NTSR1-βarr1ΔCT assembly. Receptor, GRK5 and βarr1ΔCT were purified separately. Phosphorylation of NTSR1 by GRK5 in the presence of diC8-PI(4,5)P2 was monitored by analytical IEX (shown in inset as gradient IEX elution from a representative preparation; a stepped elution IEX run is shown in Extended Data Fig. 2a), followed by M1 affinity purification. Phosphorylated receptor was combined with βarr1ΔCT and additional diC8-PI(4,5)P2 and separated by SEC to isolate NTSR1-βarr1ΔCT complex. Complex was labeled with sulfo-LC-SDA and UV irradiated. Following a second round of SEC to re-isolate complex, the sample was concentrated and used for cryo-EM. b) Representative sample preparation: phosphorylated NTSR1 was mixed with βarr1ΔCT and purified by SEC (blue curve, SEC1). Complex fractions were combined and treated with crosslinker, quenched, then UV irradiated. Lanes 2 and 3 of SDS-PAGE show pooled samples before and after crosslinking and UV irradiation; about 25% of sample is crosslinked (based on densitometry, top two bands relative to total). Crosslinked sample was re-run by SEC (red curve, SEC2) and SDS-PAGE of peak fractions is shown. Peak fractions were combined, concentrated and used for cryo-EM. Representative negative stain EM images of NTSR1-βarr1ΔCT complex show similar homogeneity, pre- and post-crosslinking. Enlarged views of rotated representative particles (denoted by yellow and blue circles) are shown. Four sample preparations, which all gave similar results by SEC, SDS-PAGE and negative stain analysis, were used to generate the cryoEM dataset.