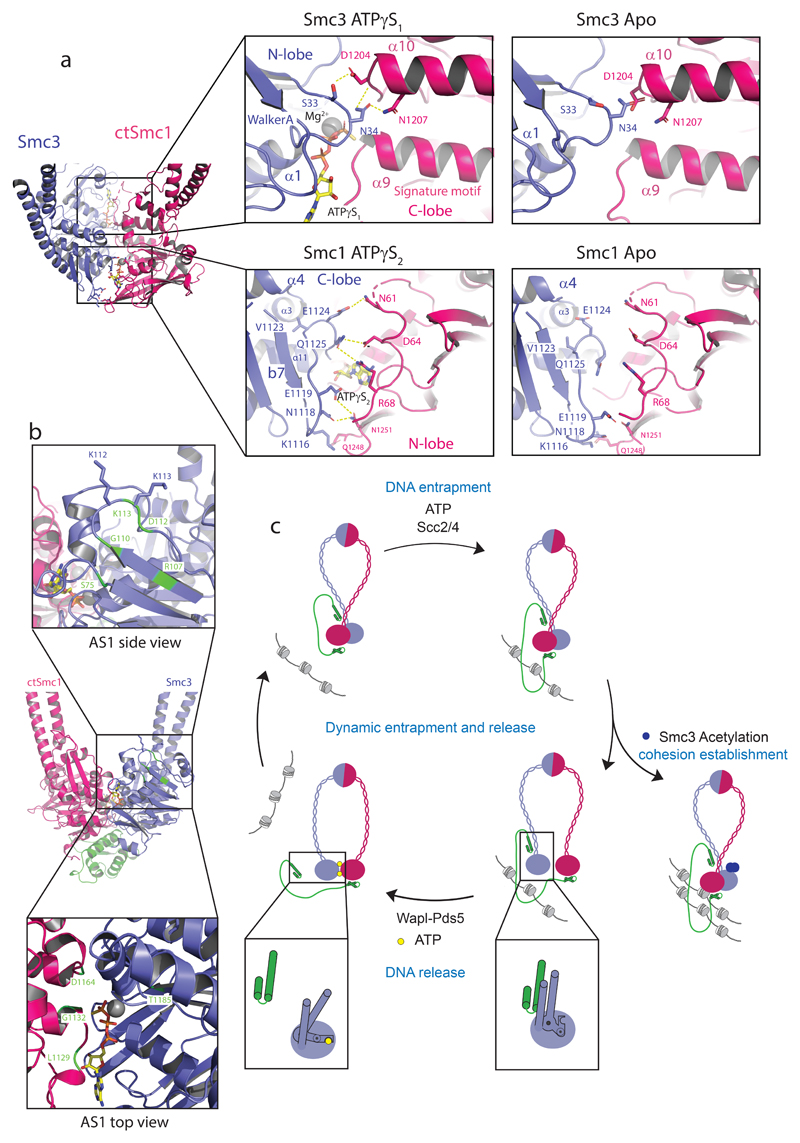
Figure 3. The Smc3 and Smc1 ATPase sites are structurally distinct.
a, Amino acid residue interactions in the ATPγS-engaged Smc3–CtSmc1 heterodimer and comparison with the hypothetical apo Smc heterodimer interface. Interactions (i.e. hydrogen bonds and salt bridges) are indicated by yellow dashed lines. b, Mutations which suppress cohesin release (green) cluster at the Smc3 ATPase site. Residues in close proximity to the Smc3 acetyl-lysines and which may influence allosteric regulation of the ATPase are depicted in the top inset. Suppressors within the Smc3 ATPase directly influence hydrolysis (bottom inset). c, Model of the cohesin ATPase cycle (Clockwise from left-hand side). DNA capture is mediated by Scc2–4 and depends on ATP. DNA release is catalyzed by Wapl–Pds5. Heterodimerization and ATP-binding causes conformational changes within the Smc3 coiled-coil domain, stabilized by a ‘latch’-like function of the Q-loop (inset) which lead to Scc1 displacement and cohesin release. ATP hydrolysis permits the cycle to resume. Cohesin is liberated from this cycle by acetylation of Smc3, which counteracts release and establishes cohesion (right-hand side).