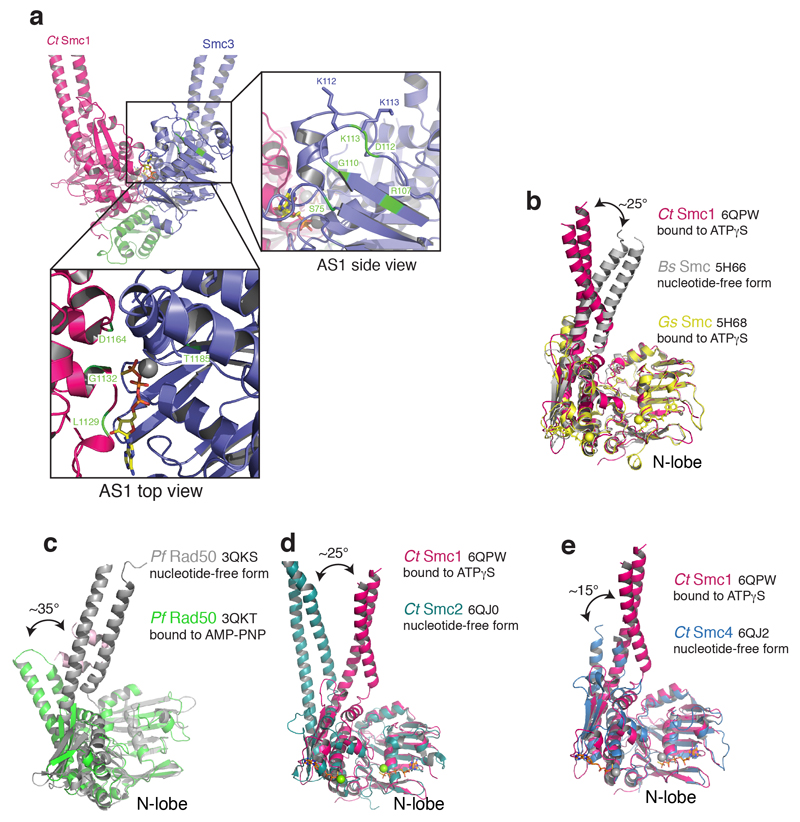
Extended Data Fig. 4. Nucleotide-induced conformational changes in SMC ATPases.
a, Structural alignment based on ATPγS-bound CtSMC1, the nucleotide-free form of Bacillus subtilis (Bs) SMC and the ATPγS-bound form of Geobacillus stearothermophilus (Gs) SMC. b, Nucleotide free (grey) and bound (green) Pyrococccus furiosis (Pf) Rad50 conformations. Nucleotide binding induces an ~35° C-lobe rotation. c, Nucleotide free (teal) form of Chaetomium thermophilium (Ct) Smc2 and ATPγS-bound form of CtSmc1 (red). d, Nucleotide free (blue) form of CtSmc4 and ATPγS-bound form of CtSmc1 (red). All structural superpositions were done using the SMC N-lobe.