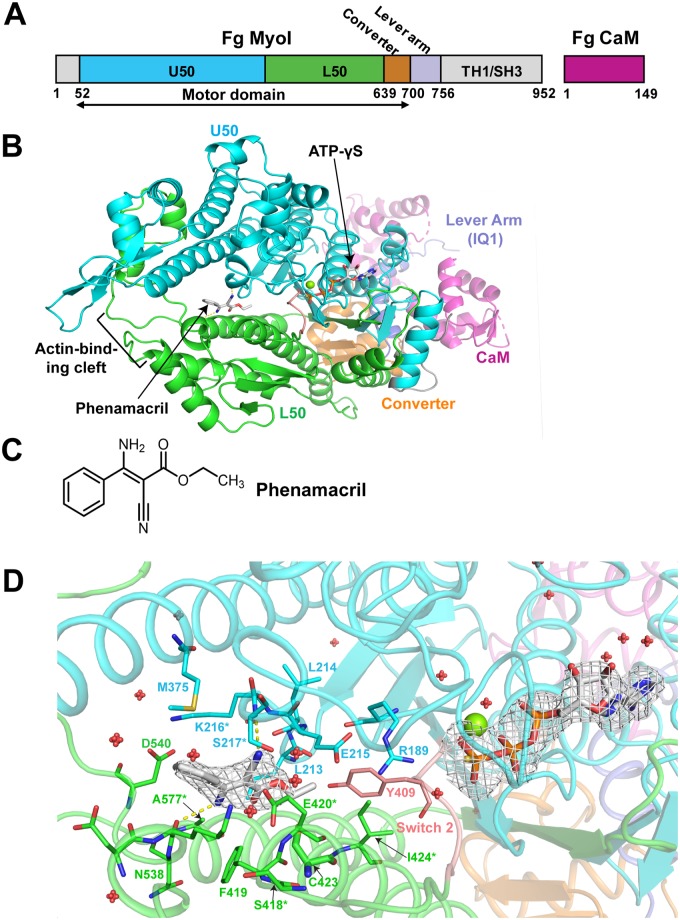
Fig 2. FgMyoI structure.
(A) Domain map of F. graminearum FgMyoI encoded by the myo5 gene in complex with F. graminearum calmodulin (CaM). (B) Structure overview. (C) Chemical structure of phenamacril. (D) Catalytic center and phenamacril pocket with ligand Fo-Fc omit map contoured at 3.0 σ. The ligand phenamacril shows strong electron density at 3 σ except the ethyl ester tail that is partially disordered due to its flexibility. Pocket residues are shown in thick line presentation, the Mg2+ ion as green sphere, and water molecules as red spheres. *: pocket residues that have been found mutated in highly resistant (K216E/R, S217L/P, E420G/K/D) or moderately resistant (S418R, I424R, A577G) Fusarium strains.